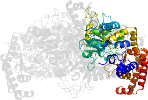
Lineage for d6smrd1 (6smr D:1-470)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.67: PLP-dependent transferase-like [53382] (3 superfamilies)
main domain: 3 layers: a/b/a, mixed beta-sheet of 7 strands, order 3245671; strand 7 is antiparallel to the rest
Superfamily c.67.1: PLP-dependent transferases [53383] (10 families) 

Family c.67.1.4: GABA-aminotransferase-like [53417] (17 proteins)
formerly omega-Aminoacid:pyruvate aminotransferase-like
Protein automated matches [190152] (25 species)
not a true protein
Species Thale cress (Arabidopsis thaliana) [TaxId:3702] [318454] (6 PDB entries) 
Domain d6smrd1: 6smr D:1-470 [378895]
Other proteins in same PDB: d6smra2, d6smrb2, d6smrc2, d6smrd2
automated match to d5v7ia_
complexed with edo, mtx, pls, trs
Details for d6smrd1
PDB Entry: 6smr (more details), 2.12 Å
PDB Description: a. thaliana serine hydroxymethyltransferase isoform 4 (atshmt4) in complex with methotrexate
PDB Compounds: (D:) Serine hydroxymethyltransferase 4SCOPe Domain Sequences for d6smrd1:
Sequence; same for both SEQRES and ATOM records: (download)
>d6smrd1 c.67.1.4 (D:1-470) automated matches {Thale cress (Arabidopsis thaliana) [TaxId: 3702]}
mepvsswgntslvsvdpeihdliekekrrqcrgieliasenftsfaviealgsaltnkys
egipgnryyggnefideienlcrsraleafhcdpaawgvnvqpysgspanfaaytallqp
hdrimgldlpsgghlthgyytsggkkisatsiyfeslpykvnfttgyidydkleekaldf
rpkllicggsayprdwdyarfraiadkvgalllcdmahisglvaaqeaanpfeycdvvtt
tthkslrgpragmifyrkgpkppkkgqpegavydfedkinfavfpalqggphnhqigala
valkqantpgfkvyakqvkanavalgnylmskgyqivtngtenhlvlwdlrplgltgnkv
eklcdlcsitlnknavfgdssalapggvrigapamtsrglvekdfeqigeflsravtltl
diqktygkllkdfnkglvnnkdldqlkadvekfsasyempgflmsemkyk
SCOPe Domain Coordinates for d6smrd1:
Click to download the PDB-style file with coordinates for d6smrd1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d6smrd1:
- d6smrd1 first appeared in SCOPe 2.07
 View in 3D
View in 3D