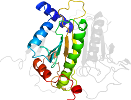
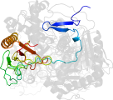
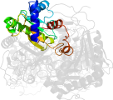
Lineage for d1gupa1 (1gup A:2-177)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.13: HIT-like [54196] (2 superfamilies)
alpha-beta(3)-alpha-beta(2); 3 layers: alpha/beta/alpha
Superfamily d.13.1: HIT-like [54197] (6 families) 
Family d.13.1.2: Hexose-1-phosphate uridylyltransferase [54207] (1 protein)
duplication: consists of 2 HIT-like motifs
binds zinc and iron ions
Protein Galactose-1-phosphate uridylyltransferase [54208] (3 species) 
Species Escherichia coli [TaxId:562] [54209] (4 PDB entries) 
Domain d1gupa1: 1gup A:2-177 [37534]
complexed with fe, gdu, k, zn
Details for d1gupa1
PDB Entry: 1gup (more details), 1.8 Å
PDB Description: structure of nucleotidyltransferase complexed with udp-galactose
PDB Compounds: (A:) galactose-1-phosphate uridylyltransferaseSCOPe Domain Sequences for d1gupa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1gupa1 d.13.1.2 (A:2-177) Galactose-1-phosphate uridylyltransferase {Escherichia coli [TaxId: 562]}
tqfnpvdhphrrynpltgqwilvsphrakrpwqgaqetpakqvlpahdpdcflcagnvrv
tgdknpdytgtyvftndfaalmsdtpdapeshdplmrcqsargtsrvicfspdhsktlpe
lsvaalteivktwqeqtaelgktypwvqvfenkgaamgcsnphpggqiwansflpn
SCOPe Domain Coordinates for d1gupa1:
Click to download the PDB-style file with coordinates for d1gupa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1gupa1:
- d1gupa1 first appeared (with stable ids) in SCOP 1.55
- d1gupa1 appears in SCOPe 2.07

 View in 3D
View in 3D