Lineage for d108la_ (108l A:)
- Root: SCOP 1.73

Class d: Alpha and beta proteins (a+b) [53931] (334 folds) 
Fold d.2: Lysozyme-like [53954] (1 superfamily)
common alpha+beta motif for the active site region
Superfamily d.2.1: Lysozyme-like [53955] (7 families) 

Family d.2.1.3: Phage lysozyme [53981] (3 proteins) 
Protein Phage T4 lysozyme [53982] (1 species) 
Species Bacteriophage T4 [TaxId:10665] [53983] (431 PDB entries)
many mutant structures
Domain d108la_: 108l A: [36669]
complexed with cl, seo; mutant
Details for d108la_
PDB Entry: 108l (more details), 1.8 Å
PDB Description: structural basis of alpha-helix propensity at two sites in t4 lysozyme
PDB Compounds: (A:) t4 lysozymeSCOP Domain Sequences for d108la_:
Sequence; same for both SEQRES and ATOM records: (download)
>d108la_ d.2.1.3 (A:) Phage T4 lysozyme {Bacteriophage T4 [TaxId: 10665]}
mnifemlrideglrlkiykdtegyytigighlltkspslnaakieldkaigrntngvitk
deaeklfnqdvdaavrgilrnaklkpvydsldavrraalinmvfqmgetgvagftnslrm
lqqkrwdeaavnlaksrwynqtpnrakrvittfrtgtwdayk
SCOP Domain Coordinates for d108la_:
Click to download the PDB-style file with coordinates for d108la_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d108la_:
- d108la_ first appeared (with stable ids) in SCOP 1.55, called d108l__
- d108la_ was called d108l__ in SCOP 1.71
- d108la_ appears in SCOP 1.75
- d108la_ appears in the current release, SCOPe 2.08
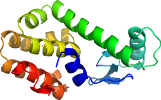
 View in 3D
View in 3D