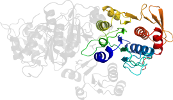
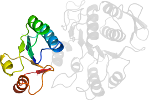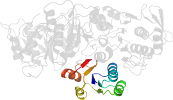Lineage for d3eaga2 (3eag A:93-323)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.72: Ribokinase-like [53612] (3 superfamilies)
core: 3 layers: a/b/a; mixed beta-sheet of 8 strands, order 21345678, strand 7 is antiparallel to the rest
potential superfamily: members of this fold have similar functions but different ATP-binding sites
Superfamily c.72.2: MurD-like peptide ligases, catalytic domain [53623] (3 families) 
has extra strand located between strands 1 and 2
Family c.72.2.0: automated matches [254328] (1 protein)
not a true family
Protein automated matches [254749] (4 species)
not a true protein
Species Neisseria meningitidis [TaxId:122586] [346327] (1 PDB entry) 
Domain d3eaga2: 3eag A:93-323 [344705]
Other proteins in same PDB: d3eaga1, d3eaga3, d3eagb1, d3eagb3
automated match to d5vvwa2
complexed with gol
Details for d3eaga2
PDB Entry: 3eag (more details), 2.55 Å
PDB Description: The crystal structure of UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase (MPL) from Neisseria meningitides
PDB Compounds: (A:) UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligaseSCOPe Domain Sequences for d3eaga2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3eaga2 c.72.2.0 (A:93-323) automated matches {Neisseria meningitidis [TaxId: 122586]}
gpqwlsenvlhhhwvlgvagthgktttasmlawvleyaglapgfliggvpenfgvsarlp
qtprqdpnsqspffvieadeydtaffdkrskfvhyrprtavlnnlefdhadifadlgaiq
tqfhylvrtvpseglivcngrqqslqdtldkgcwtpvekfgtehgwqageanadgsfdvl
ldgktagrvkwdlmgrhnrmnalaviaaarhvgvdiqtacealgafknvkr
SCOPe Domain Coordinates for d3eaga2:
Click to download the PDB-style file with coordinates for d3eaga2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3eaga2:
- d3eaga2 first appeared in SCOPe 2.07
 View in 3D
View in 3D