Lineage for d1dkoa_ (1dko A:)
- Root: SCOPe 2.05

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.60: Phosphoglycerate mutase-like [53253] (1 superfamily)
core: 3 layers, a/b/a; mixed beta-sheet of 6 strands, order 324156; strand 5 is antiparallel to the rest
Superfamily c.60.1: Phosphoglycerate mutase-like [53254] (4 families) 

Family c.60.1.2: Histidine acid phosphatase [53258] (4 proteins) 
Protein Phytase (myo-inositol-hexakisphosphate-3-phosphohydrolase) [53263] (4 species) 
Species Escherichia coli [TaxId:562] [53266] (6 PDB entries) 
Domain d1dkoa_: 1dko A: [34001]
complexed with hg, wo4
Details for d1dkoa_
PDB Entry: 1dko (more details), 2.38 Å
PDB Description: crystal structure of tungstate complex of escherichia coli phytase at ph 6.6 with tungstate bound at the active site and with hg2+ cation acting as an intermolecular bridge
PDB Compounds: (A:) phytaseSCOPe Domain Sequences for d1dkoa_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1dkoa_ c.60.1.2 (A:) Phytase (myo-inositol-hexakisphosphate-3-phosphohydrolase) {Escherichia coli [TaxId: 562]}
qsepelklesvvivsrhgvraptkatqlmqdvtpdawptwpvklgwltprggeliaylgh
yqrqrlvadgllakkgcpqsgqvaiiadvdertrktgeafaaglapdcaitvhtqtdtss
pdplfnplktgvcqldnanvtdailsraggsiadftghrqtafrelervlnfpqsnlclk
rekqdescsltqalpselkvsadnvsltgavslasmlteifllqqaqgmpepgwgritds
hqwntllslhnaqfyllqrtpevarsratplldliktaltphppqkqaygvtlptsvlfi
aghdtnlanlggalelnwtlpgqpdntppggelvferwrrlsdnsqwiqvslvfqtlqqm
rdktplslntppgevkltlagceernaqgmcslagftqivnearipacsl
SCOPe Domain Coordinates for d1dkoa_:
Click to download the PDB-style file with coordinates for d1dkoa_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1dkoa_:
- d1dkoa_ first appeared (with stable ids) in SCOP 1.55
- d1dkoa_ appears in SCOPe 2.04
- d1dkoa_ appears in SCOPe 2.06
- d1dkoa_ appears in the current release, SCOPe 2.08
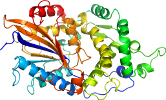
 View in 3D
View in 3D