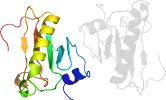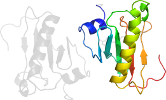Lineage for d5uvma1 (5uvm A:2-114)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.13: HIT-like [54196] (2 superfamilies)
alpha-beta(3)-alpha-beta(2); 3 layers: alpha/beta/alpha
Superfamily d.13.1: HIT-like [54197] (6 families) 

Family d.13.1.1: HIT (HINT, histidine triad) family of protein kinase-interacting proteins [54198] (8 proteins)
Pfam PF01230
topologically similar to the N-terminal domain of protein kinases
Protein Putative hydrolase [117791] (1 species) 
Species Ruminiclostridium thermocellum [TaxId:637887] [332789] (1 PDB entry) 
Domain d5uvma1: 5uvm A:2-114 [332860]
Other proteins in same PDB: d5uvma2, d5uvmb2
automated match to d1xqub_
complexed with adn, unx, zn
Details for d5uvma1
PDB Entry: 5uvm (more details), 2.3 Å
PDB Description: hit family hydrolase from clostridium thermocellum cth-393
PDB Compounds: (A:) Histidine triad (HIT) proteinSCOPe Domain Sequences for d5uvma1:
Sequence; same for both SEQRES and ATOM records: (download)
>d5uvma1 d.13.1.1 (A:2-114) Putative hydrolase {Ruminiclostridium thermocellum [TaxId: 637887]}
encvfckiikrelpstiyyederviaikdinpaapvhvliipkehianvkeinesnaqil
idihkaankvaedlgiaekgyrlitncgvaagqtvfhlhyhllggvdmgpkil
SCOPe Domain Coordinates for d5uvma1:
Click to download the PDB-style file with coordinates for d5uvma1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d5uvma1:
- d5uvma1 first appeared in SCOPe 2.06
- d5uvma1 appears in SCOPe 2.07
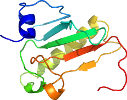
 View in 3D
View in 3D