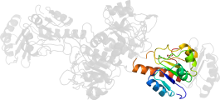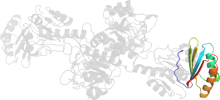Lineage for d1psda2 (1psd A:7-107,A:296-326)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.23: Flavodoxin-like [52171] (15 superfamilies)
3 layers, a/b/a; parallel beta-sheet of 5 strand, order 21345
Superfamily c.23.12: Formate/glycerate dehydrogenase catalytic domain-like [52283] (3 families) 

Family c.23.12.1: Formate/glycerate dehydrogenases, substrate-binding domain [52284] (7 proteins)
this domain is interrupted by the Rossmann-fold domain
Protein Phosphoglycerate dehydrogenase [52293] (2 species)
has additional C-terminal domain of the ferredoxin fold
Species Escherichia coli [TaxId:562] [52294] (7 PDB entries) 
Domain d1psda2: 1psd A:7-107,A:296-326 [31357]
Other proteins in same PDB: d1psda1, d1psda3, d1psdb1, d1psdb3
complexed with nad, ser
Details for d1psda2
PDB Entry: 1psd (more details), 2.75 Å
PDB Description: the allosteric ligand site in the vmax-type cooperative enzyme phosphoglycerate dehydrogenase
PDB Compounds: (A:) d-3-phosphoglycerate dehydrogenase (phosphoglycerate dehydrogenase)SCOPe Domain Sequences for d1psda2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1psda2 c.23.12.1 (A:7-107,A:296-326) Phosphoglycerate dehydrogenase {Escherichia coli [TaxId: 562]}
ekdkikfllvegvhqkaleslraagytniefhkgalddeqlkesirdahfiglrsrthlt
edvinaaeklvaigcfcigtnqvdldaaakrgipvfnapfsXstqeaqeniglevagkli
kysdngstlsavn
SCOPe Domain Coordinates for d1psda2:
Click to download the PDB-style file with coordinates for d1psda2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1psda2:
- d1psda2 first appeared (with stable ids) in SCOP 1.55
- d1psda2 appears in SCOPe 2.07

 View in 3D
View in 3D