Lineage for d4x2wa_ (4x2w A:)
- Root: SCOPe 2.08

Class b: All beta proteins [48724] (180 folds) 
Fold b.47: Trypsin-like serine proteases [50493] (1 superfamily)
barrel, closed; n=6, S=8; greek-key
duplication: consists of two domains of the same fold
Superfamily b.47.1: Trypsin-like serine proteases [50494] (5 families) 
Family b.47.1.4: Viral cysteine protease of trypsin fold [50603] (5 proteins) 
Protein automated matches [190384] (21 species)
not a true protein
Species Murine norovirus 1 [TaxId:223997] [311357] (5 PDB entries) 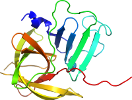
Domain d4x2wa_: 4x2w A: [309951]
automated match to d2fyqa_
mutant
Details for d4x2wa_
PDB Entry: 4x2w (more details), 2.7 Å
PDB Description: crystal structure of the murine norovirus ns6 protease (inactive c139a mutant) with a c-terminal extension to include residues p1 prime - p2 prime of ns7
PDB Compounds: (A:) ns6 proteaseSCOPe Domain Sequences for d4x2wa_:
Sequence, based on SEQRES records: (download)
>d4x2wa_ b.47.1.4 (A:) automated matches {Murine norovirus 1 [TaxId: 223997]}
vsiwsrvvqfgtgwgfwvsghvfitakhvappkgteifgrkpgdftvtssgdflkyyfts
avrpdipamvlengcqegvvasvlvkrasgemlalavrmgsqaaikigsavvhgqtgmll
tgsnakaqdlgtipgdagcpyvykkgntwvvigvhvaatrsgntviaathgeptleale
Sequence, based on observed residues (ATOM records): (download)
>d4x2wa_ b.47.1.4 (A:) automated matches {Murine norovirus 1 [TaxId: 223997]}
vsiwsrvvqfgtgwgfwvsghvfitakhvappkgteifgrkpgdftvtssgdflkyyfts
avrpdipamvlengcqegvvasvlvkrasgemlalavrmgsqaaikigsavvhgqtgmll
tqdlgtipgdagcpyvykkgntwvvigvhvaatrsgntviaathgeptleale
SCOPe Domain Coordinates for d4x2wa_:
Click to download the PDB-style file with coordinates for d4x2wa_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4x2wa_:
- d4x2wa_ first appeared in SCOPe 2.06
- d4x2wa_ appears in SCOPe 2.07
 View in 3D
View in 3D
