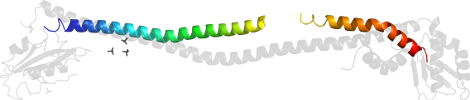
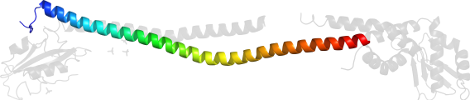Lineage for d4efag_ (4efa G:)
- Root: SCOPe 2.06

Class h: Coiled coil proteins [57942] (7 folds) 
Fold h.1: Parallel coiled-coil [57943] (38 superfamilies)
this is not a true fold; includes oligomers of shorter identical helices
Superfamily h.1.37: V-type ATPase peripheral stalk subunit G coiled coil [310579] (1 family) 
Unusual right-handed coiled coil noted in PubMed 20173764
Family h.1.37.1: V-type ATPase peripheral stalk subunit G coiled coil [310619] (2 proteins) 
Protein automated matches [310885] (1 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:559292] [311372] (2 PDB entries) 
Domain d4efag_: 4efa G: [307296]
Other proteins in same PDB: d4efae1, d4efae2
automated match to d3j9th_
complexed with so4
Details for d4efag_
PDB Entry: 4efa (more details), 2.82 Å
PDB Description: crystal structure of the heterotrimeric egchead peripheral stalk complex of the yeast vacuolar atpase - second conformation
PDB Compounds: (G:) V-type proton ATPase subunit GSCOPe Domain Sequences for d4efag_:
Sequence, based on SEQRES records: (download)
>d4efag_ h.1.37.1 (G:) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 559292]}
sqkngiatllqaekeaheivskarkyrqdklkqaktdaakeidsykiqkdkelkefeqkn
aggvgelekkaeagvqgelaeikkiaekkkddvvkilietvikp
Sequence, based on observed residues (ATOM records): (download)
>d4efag_ h.1.37.1 (G:) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 559292]}
sqkngiatllqaekeaheivskarkyrqdklkqaktdaakeidsykiqkdkelkefeqkn
kaeagvqgelaeikkiaekkkddvvkilietvikp
SCOPe Domain Coordinates for d4efag_:
Click to download the PDB-style file with coordinates for d4efag_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4efag_:
- d4efag_ is new in SCOPe 2.06-stable
- d4efag_ appears in SCOPe 2.07
- d4efag_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D