Lineage for d1hyhc1 (1hyh C:21-166)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.2: NAD(P)-binding Rossmann-fold domains [51734] (1 superfamily)
core: 3 layers, a/b/a; parallel beta-sheet of 6 strands, order 321456
The nucleotide-binding modes of this and the next two folds/superfamilies are similar
Superfamily c.2.1: NAD(P)-binding Rossmann-fold domains [51735] (13 families) 

Family c.2.1.5: LDH N-terminal domain-like [51848] (9 proteins) 
Protein L-2-hydroxyisocapronate dehydrogenase, L-HICDH [51857] (1 species) 
Species Lactobacillus confusus [TaxId:1583] [51858] (1 PDB entry) 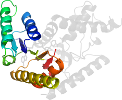
Domain d1hyhc1: 1hyh C:21-166 [30152]
Other proteins in same PDB: d1hyha2, d1hyhb2, d1hyhc2, d1hyhd2
complexed with nad, so4
Details for d1hyhc1
PDB Entry: 1hyh (more details), 2.2 Å
PDB Description: crystal structure of l-2-hydroxyisocaproate dehydrogenase from lactobacillus confusus at 2.2 angstroms resolution-an example of strong asymmetry between subunits
PDB Compounds: (C:) l-2-hydroxyisocaproate dehydrogenaseSCOPe Domain Sequences for d1hyhc1:
Sequence, based on SEQRES records: (download)
>d1hyhc1 c.2.1.5 (C:21-166) L-2-hydroxyisocapronate dehydrogenase, L-HICDH {Lactobacillus confusus [TaxId: 1583]}
arkigiiglgnvgaavahgliaqgvaddyvfidaneakvkadqidfqdamanleahgniv
indwaaladadvvistlgniklqqdnptgdrfaelkftssmvqsvgtnlkesgfhgvlvv
isnpvdvitalfqhvtgfpahkvigt
Sequence, based on observed residues (ATOM records): (download)
>d1hyhc1 c.2.1.5 (C:21-166) L-2-hydroxyisocapronate dehydrogenase, L-HICDH {Lactobacillus confusus [TaxId: 1583]}
arkigiiglgnvgaavahgliaqgvaddyvfidaneakvkadqidfqdamanleahgniv
indwaaladadvvistlggdrfaelkftssmvqsvgtnlkesgfhgvlvvisnpvdvita
lfqhvtgfpahkvigt
SCOPe Domain Coordinates for d1hyhc1:
Click to download the PDB-style file with coordinates for d1hyhc1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1hyhc1:
- d1hyhc1 first appeared (with stable ids) in SCOP 1.55
- d1hyhc1 appears in SCOPe 2.07
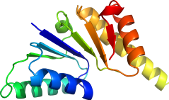
 View in 3D
View in 3D