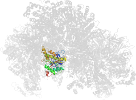
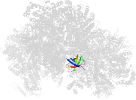
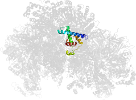
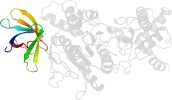Lineage for d3oehv3 (3oeh V:358-475)
- Root: SCOPe 2.05

Class a: All alpha proteins [46456] (286 folds) 
Fold a.69: Left-handed superhelix [47916] (4 superfamilies)
core: 4-5 helices; bundle; left-handed superhelix
Superfamily a.69.1: C-terminal domain of alpha and beta subunits of F1 ATP synthase [47917] (2 families) 

Family a.69.1.0: automated matches [254233] (1 protein)
not a true family
Protein automated matches [254528] (7 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [255214] (6 PDB entries) 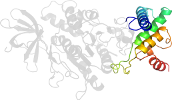
Domain d3oehv3: 3oeh V:358-475 [248239]
Other proteins in same PDB: d3oehd1, d3oehd2, d3oehe1, d3oehe2, d3oehf1, d3oehf2, d3oehg_, d3oehm1, d3oehm2, d3oehn1, d3oehn2, d3oeho1, d3oeho2, d3oehp_, d3oehv1, d3oehv2, d3oehw1, d3oehw2, d3oehx1, d3oehx2
automated match to d2jdid1
complexed with anp, mg; mutant
Details for d3oehv3
PDB Entry: 3oeh (more details), 3 Å
PDB Description: Structure of four mutant forms of yeast F1 ATPase: beta-V279F
PDB Compounds: (V:) ATP synthase subunit betaSCOPe Domain Sequences for d3oehv3:
Sequence; same for both SEQRES and ATOM records: (download)
>d3oehv3 a.69.1.0 (V:358-475) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
ldaavvgqehydvaskvqetlqtykslqdiiailgmdelseqdkltverarkiqrflsqp
favaevftgipgklvrlkdtvasfkavlegkydnipehafymvggiedvvakaeklaa
SCOPe Domain Coordinates for d3oehv3:
Click to download the PDB-style file with coordinates for d3oehv3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3oehv3:
- d3oehv3 first appeared in SCOPe 2.04
- d3oehv3 appears in SCOPe 2.06
- d3oehv3 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d3oehd1, d3oehd2, d3oehd3, d3oehe1, d3oehe2, d3oehe3, d3oehf1, d3oehf2, d3oehf3, d3oehg_, d3oehm1, d3oehm2, d3oehm3, d3oehn1, d3oehn2, d3oehn3, d3oeho1, d3oeho2, d3oeho3, d3oehp_, d3oehw1, d3oehw2, d3oehw3, d3oehx1, d3oehx2, d3oehx3 |