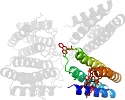
Lineage for d3nmjb_ (3nmj B:)
- Root: SCOPe 2.05

Class a: All alpha proteins [46456] (286 folds) 
Fold a.24: Four-helical up-and-down bundle [47161] (28 superfamilies)
core: 4 helices; bundle, closed or partly opened, left-handed twist; up-and-down
Superfamily a.24.3: Cytochromes [47175] (3 families) 
Heme-containing proteins
Family a.24.3.1: Cytochrome b562 [47176] (2 proteins)
automatically mapped to Pfam PF07361
Protein automated matches [190502] (2 species)
not a true protein
Species Escherichia coli [TaxId:562] [187450] (34 PDB entries) 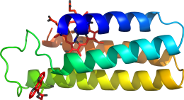
Domain d3nmjb_: 3nmj B: [247961]
automated match to d3nmia_
complexed with hem, ni, pxx
Details for d3nmjb_
PDB Entry: 3nmj (more details), 3.1 Å
PDB Description: Crystal structure of a nickel mediated dimer for the phenanthroline-modified cytochrome cb562 variant, MBP-Phen2
PDB Compounds: (B:) Soluble cytochrome b562SCOPe Domain Sequences for d3nmjb_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3nmjb_ a.24.3.1 (B:) automated matches {Escherichia coli [TaxId: 562]}
dlednmetlndnlkviekadnaaqvkdaltkmraaaldaqkatppkledkspdspemcdf
aagfailvgqiddalklanegkvkeaqaaaeqlkttcnachqkyr
SCOPe Domain Coordinates for d3nmjb_:
Click to download the PDB-style file with coordinates for d3nmjb_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3nmjb_:
- d3nmjb_ first appeared in SCOPe 2.04
- d3nmjb_ appears in SCOPe 2.06
- d3nmjb_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D