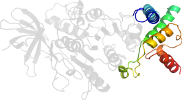Lineage for d3ziap2 (3zia P:83-357)
- Root: SCOPe 2.04

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (25 families) 
division into families based on beta-sheet topologies
Family c.37.1.11: RecA protein-like (ATPase-domain) [52670] (18 proteins)
core: mixed beta-sheet of 8 strands, order 32451678; strand 7 is antiparallel to the rest
Protein automated matches [190393] (9 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [255213] (6 PDB entries) 
Domain d3ziap2: 3zia P:83-357 [239941]
Other proteins in same PDB: d3ziad1, d3ziad3, d3ziae1, d3ziae3, d3ziaf1, d3ziaf3, d3ziag_, d3zian1, d3zian3, d3ziao1, d3ziao3, d3ziap1, d3ziap3, d3ziaq_
automated match to d2jdid3
complexed with adp, atp, edo, mg
Details for d3ziap2
PDB Entry: 3zia (more details), 2.5 Å
PDB Description: the structure of f1-atpase from saccharomyces cerevisiae inhibited by its regulatory protein if1
PDB Compounds: (P:) ATP synthase subunit beta, mitochondrialSCOPe Domain Sequences for d3ziap2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3ziap2 c.37.1.11 (P:83-357) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
isvpvgretlgriinvigepidergpiksklrkpihadppsfaeqstsaeiletgikvvd
llapyarggkiglfggagvgktvfiqelinniakahggfsvftgvgertregndlyremk
etgvinlegeskvalvfgqmneppgararvaltgltiaeyfrdeegqdvllfidnifrft
qagsevsallgripsavgyqptlatdmgllqeritttkkgsvtsvqavyvpaddltdpap
attfahldattvlsrgiselgiypavdpldsksrl
SCOPe Domain Coordinates for d3ziap2:
Click to download the PDB-style file with coordinates for d3ziap2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3ziap2:
- d3ziap2 is new in SCOPe 2.04-stable
- d3ziap2 appears in SCOPe 2.05
- d3ziap2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D