Lineage for d4ivfb2 (4ivf B:95-222)
- Root: SCOPe 2.05

Class a: All alpha proteins [46456] (286 folds) 
Fold a.45: GST C-terminal domain-like [47615] (1 superfamily)
core: 4 helices; bundle, closed, left-handed twist; right-handed superhelix
Superfamily a.45.1: GST C-terminal domain-like [47616] (3 families) 
this domains follows the thioredoxin-like N-terminal domain
Family a.45.1.0: automated matches [227130] (1 protein)
not a true family
Protein automated matches [226831] (51 species)
not a true protein
Species Lodderomyces elongisporus [TaxId:379508] [234908] (1 PDB entry) 
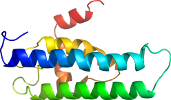
Domain d4ivfb2: 4ivf B:95-222 [234909]
Other proteins in same PDB: d4ivfa1, d4ivfb1, d4ivfc1, d4ivfd1, d4ivfe1, d4ivff1, d4ivfg1, d4ivfh1
automated match to d4ecib2
complexed with cit, gsh
Details for d4ivfb2
PDB Entry: 4ivf (more details), 2.2 Å
PDB Description: crystal structure of glutathione transferase homolog from lodderomyces elongisporus, target efi-501753, with two gsh per subunit
PDB Compounds: (B:) Putative uncharacterized proteinSCOPe Domain Sequences for d4ivfb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d4ivfb2 a.45.1.0 (B:95-222) automated matches {Lodderomyces elongisporus [TaxId: 379508]}
fsypagtaeyyktleylifqvaengpiqgqanhfvfaakekvpyginryitdtkriygvf
edilsrnkandskylvgdrytvadfallgwayrlsrleidinqwpllgkwydsllklpav
qkgfevpp
SCOPe Domain Coordinates for d4ivfb2:
Click to download the PDB-style file with coordinates for d4ivfb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4ivfb2:
- d4ivfb2 first appeared in SCOPe 2.03
- d4ivfb2 appears in SCOPe 2.04
- d4ivfb2 appears in SCOPe 2.06
- d4ivfb2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D