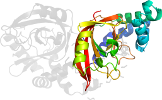
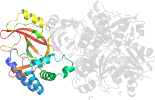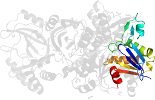Lineage for d4h0va1 (4h0v A:1-209)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.166: ADP-ribosylation [56398] (1 superfamily)
unusual fold
Superfamily d.166.1: ADP-ribosylation [56399] (8 families) 
Family d.166.1.1: ADP-ribosylating toxins [56400] (10 proteins) 
Protein automated matches [190133] (8 species)
not a true protein
Species Clostridium perfringens [TaxId:1502] [234554] (8 PDB entries) 
Domain d4h0va1: 4h0v A:1-209 [234593]
Other proteins in same PDB: d4h0vb1, d4h0vb2
automated match to d1giqa1
complexed with atp, ca, edo, lar, nad, po4
Details for d4h0va1
PDB Entry: 4h0v (more details), 2.03 Å
PDB Description: Crystal structure of NAD+-Ia(E378S)-actin complex
PDB Compounds: (A:) Iota toxin component IaSCOPe Domain Sequences for d4h0va1:
Sequence; same for both SEQRES and ATOM records: (download)
>d4h0va1 d.166.1.1 (A:1-209) automated matches {Clostridium perfringens [TaxId: 1502]}
afierpedflkdkenaiqwekkeaerveknldtlekealelykkdseqisnysqtrqyfy
dyqiesnprekeyknlrnaisknkidkpinvyyfespekfafnkeirtenqneislekfn
elketiqdklfkqdgfkdvslyepgngdekptpllihlklpkntgmlpyinsndvktlie
qdysikidkivriviegkqyikaeasivn
SCOPe Domain Coordinates for d4h0va1:
Click to download the PDB-style file with coordinates for d4h0va1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4h0va1:
- d4h0va1 first appeared in SCOPe 2.03
- d4h0va1 appears in SCOPe 2.07
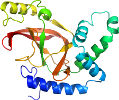
 View in 3D
View in 3D