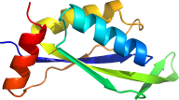
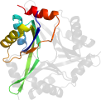
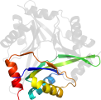
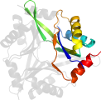
Lineage for d4iyqa1 (4iyq A:2-107)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.58: Ferredoxin-like [54861] (62 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.5: GlnB-like [54913] (6 families) 
form timeric structures with the orthogonally packed beta-sheets
Family d.58.5.0: automated matches [191474] (1 protein)
not a true family
Protein automated matches [190753] (21 species)
not a true protein
Species Ehrlichia chaffeensis [TaxId:205920] [226594] (1 PDB entry) 
Domain d4iyqa1: 4iyq A:2-107 [223481]
Other proteins in same PDB: d4iyqa2, d4iyqb2, d4iyqc2
automated match to d2zfha1
complexed with ca
Details for d4iyqa1
PDB Entry: 4iyq (more details), 2.55 Å
PDB Description: Crystal structure of divalent ion tolerance protein CutA1 from Ehrlichia chaffeensis
PDB Compounds: (A:) Divalent ion tolerance protein CutA1SCOPe Domain Sequences for d4iyqa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d4iyqa1 d.58.5.0 (A:2-107) automated matches {Ehrlichia chaffeensis [TaxId: 205920]}
mknisllytttptyedayrisnillenkliacanifsnitsvyvwedeihnntecaiilk
ttndlvqhatnkiqaihpydtpaiitidptnandkfiqwvndctal
SCOPe Domain Coordinates for d4iyqa1:
Click to download the PDB-style file with coordinates for d4iyqa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4iyqa1:
- d4iyqa1 first appeared in SCOPe 2.03, called d4iyqa_
- d4iyqa1 appears in SCOPe 2.07
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d4iyqb1, d4iyqb2, d4iyqc1, d4iyqc2 |