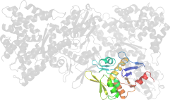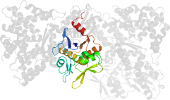Lineage for d3qmlb1 (3qml B:50-232)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.55: Ribonuclease H-like motif [53066] (7 superfamilies)
3 layers: a/b/a; mixed beta-sheet of 5 strands, order 32145; strand 2 is antiparallel to the rest
Superfamily c.55.1: Actin-like ATPase domain [53067] (16 families) 
duplication contains two domains of this fold
Family c.55.1.0: automated matches [227137] (1 protein)
not a true family
Protein automated matches [226839] (63 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [225411] (5 PDB entries) 
Domain d3qmlb1: 3qml B:50-232 [215298]
automated match to d1qqma1
complexed with mg, po4
Details for d3qmlb1
PDB Entry: 3qml (more details), 2.31 Å
PDB Description: The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor
PDB Compounds: (B:) 78 kDa glucose-regulated protein homologSCOPe Domain Sequences for d3qmlb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d3qmlb1 c.55.1.0 (B:50-232) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
gtvigidlgttyscvavmkngkteilaneqgnritpsyvaftdderligdaaknqvaanp
qntifdikrliglkyndrsvqkdikhlpfnvvnkdgkpavevsvkgekkvftpeeisgmi
lgkmkqiaedylgtkvthavvtvpayfndaqrqatkdagtiaglnvlrivneptaaaiay
gld
SCOPe Domain Coordinates for d3qmlb1:
Click to download the PDB-style file with coordinates for d3qmlb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3qmlb1:
- d3qmlb1 first appeared in SCOPe 2.03
- d3qmlb1 appears in SCOPe 2.06
- d3qmlb1 appears in the current release, SCOPe 2.08
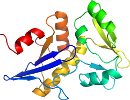
 View in 3D
View in 3D