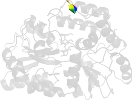Lineage for d3hstc1 (3hst C:10-374)
- Root: SCOPe 2.06

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.94: Periplasmic binding protein-like II [53849] (1 superfamily)
consists of two similar intertwined domain with 3 layers (a/b/a) each: duplication
mixed beta-sheet of 5 strands, order 21354; strand 5 is antiparallel to the rest
Superfamily c.94.1: Periplasmic binding protein-like II [53850] (4 families) 
Similar in architecture to the superfamily I but partly differs in topology
Family c.94.1.1: Phosphate binding protein-like [53851] (45 proteins) 
Protein automated matches [190140] (30 species)
not a true protein
Species Escherichia coli K-12 [TaxId:83333] [189211] (10 PDB entries) 
Domain d3hstc1: 3hst C:10-374 [211303]
Other proteins in same PDB: d3hsta2, d3hstc2
automated match to d1laxa_
complexed with edo, mlr, tar
Details for d3hstc1
PDB Entry: 3hst (more details), 2.25 Å
PDB Description: n-terminal rnase h domain of rv2228c from mycobacterium tuberculosis as a fusion protein with maltose binding protein
PDB Compounds: (C:) Maltose-binding periplasmic proteinSCOPe Domain Sequences for d3hstc1:
Sequence; same for both SEQRES and ATOM records: (download)
>d3hstc1 c.94.1.1 (C:10-374) automated matches {Escherichia coli K-12 [TaxId: 83333]}
ieegklviwingdkgynglaevgkkfekdtgikvtvehpdkleekfpqvaatgdgpdiif
wahdrfggyaqsgllaeitpdkafqdklypftwdavryngkliaypiavealsliynkdl
lpnppktweeipaldkelkakgksalmfnlqepyftwpliaadggyafkyengkydikdv
gvdnagakagltflvdliknkhmnadtdysiaeaafnkgetamtingpwawsnidtskvn
ygvtvlptfkgqpskpfvgvlsaginaaspnkelakeflenylltdegleavnkdkplga
valksyeeelakdpriaatmenaqkgeimpnipqmsafwyavrtavinaasgrqtvdeal
kdaqt
SCOPe Domain Coordinates for d3hstc1:
Click to download the PDB-style file with coordinates for d3hstc1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3hstc1:
- d3hstc1 first appeared in SCOPe 2.03, called d3hstc_
- d3hstc1 was called d3hstc_ in SCOPe 2.05
- d3hstc1 appears in SCOPe 2.07
- d3hstc1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D