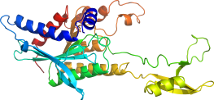
Lineage for d3h0sc1 (3h0s C:1492-1814)
- Root: SCOPe 2.05

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.14: ClpP/crotonase [52095] (1 superfamily)
core: 4 turns of (beta-beta-alpha)n superhelix
Superfamily c.14.1: ClpP/crotonase [52096] (5 families) 

Family c.14.1.4: Biotin dependent carboxylase carboxyltransferase domain [89572] (7 proteins)
Pfam PF01039
the active site is formed by two different homologous subunits or domains of this fold
Protein automated matches [226926] (3 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [225864] (7 PDB entries) 
Domain d3h0sc1: 3h0s C:1492-1814 [210774]
automated match to d1uyrb1
complexed with b38, so4
Details for d3h0sc1
PDB Entry: 3h0s (more details), 2.43 Å
PDB Description: crystal structure of the carboxyltransferase domain of acetyl-coenzyme a carboxylase in complex with compound 7
PDB Compounds: (C:) acetyl-coa carboxylaseSCOPe Domain Sequences for d3h0sc1:
Sequence; same for both SEQRES and ATOM records: (download)
>d3h0sc1 c.14.1.4 (C:1492-1814) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
wlqpkrykahlmgttyvydfpelfrqasssqwknfsadvkltddffisneliedengelt
everepganaigmvafkitvktpeyprgrqfvvvanditfkigsfgpqedeffnkvteya
rkrgipriylaansgarigmaeeivplfqvawndaanpdkgfqylyltsegmetlkkfdk
ensvltertvingeerfviktiigsedglgveclrgsgliagatsrayhdiftitlvtcr
svgigaylvrlgqraiqvegqpiiltgapainkmlgrevytsnlqlggtqimynngvshl
tavddlagvekivewmsyvpakr
SCOPe Domain Coordinates for d3h0sc1:
Click to download the PDB-style file with coordinates for d3h0sc1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3h0sc1:
- d3h0sc1 first appeared in SCOPe 2.03
- d3h0sc1 appears in SCOPe 2.04
- d3h0sc1 appears in SCOPe 2.06
- d3h0sc1 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d3h0sa1, d3h0sa2, d3h0sb1, d3h0sb2 |