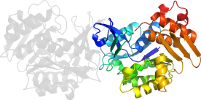Lineage for d3ewma_ (3ewm A:)
- Root: SCOPe 2.05

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.72: Ribokinase-like [53612] (3 superfamilies)
core: 3 layers: a/b/a; mixed beta-sheet of 8 strands, order 21345678, strand 7 is antiparallel to the rest
potential superfamily: members of this fold have similar functions but different ATP-binding sites
Superfamily c.72.1: Ribokinase-like [53613] (6 families) 
has extra strand located between strands 2 and 3
Family c.72.1.0: automated matches [191321] (1 protein)
not a true family
Protein automated matches [190117] (37 species)
not a true protein
Species Pyrococcus horikoshii [TaxId:53953] [225557] (2 PDB entries) 
Domain d3ewma_: 3ewm A: [209687]
automated match to d2fv7a1
Details for d3ewma_
PDB Entry: 3ewm (more details), 1.9 Å
PDB Description: crystal structure of an uncharacterized sugar kinase ph1459 from pyrococcus horikoshii
PDB Compounds: (A:) Uncharacterized sugar kinase PH1459SCOPe Domain Sequences for d3ewma_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3ewma_ c.72.1.0 (A:) automated matches {Pyrococcus horikoshii [TaxId: 53953]}
sliasigellidlisveegdlkdvrlfekhpggapanvavgvsrlgvkssliskvgndpf
geylieelskenvdtrgivkdekkhtgivfvqlkgaspsfllyddvayfnmtlndinwdi
veeakivnfgsvilarnpsretvmkvikkikgssliafdvnlrldlwrgqeeemikvlee
sikladivkaseeevlylenqgvevkgsmltaitlgpkgcrliknetvvdvpsynvnpld
ttgagdafmaallvgilklkgldllklgkfanlvaalstqkrgawstprkdellkykear
evla
SCOPe Domain Coordinates for d3ewma_:
Click to download the PDB-style file with coordinates for d3ewma_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3ewma_:
- d3ewma_ first appeared in SCOPe 2.03
- d3ewma_ appears in SCOPe 2.04
- d3ewma_ is called d3ewma1 in SCOPe 2.06
- d3ewma_ appears in the current release, SCOPe 2.08, called d3ewma1
 View in 3D
View in 3D