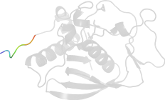Lineage for d3cmda1 (3cmd A:1-187)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.167: Peptide deformylase [56419] (1 superfamily)
alpha-beta(5)-alpha; 3 layers: a/b/a; meander beta-sheet wraps around the C-terminal alpha-helix
Superfamily d.167.1: Peptide deformylase [56420] (2 families) 
nickel-dependent enzyme
Family d.167.1.1: Peptide deformylase [56421] (2 proteins)
automatically mapped to Pfam PF01327
Protein automated matches [190200] (10 species)
not a true protein
Species Enterococcus faecium [TaxId:1352] [226815] (1 PDB entry) 
Domain d3cmda1: 3cmd A:1-187 [208899]
Other proteins in same PDB: d3cmda2, d3cmdb2
automated match to d2oklb_
complexed with fe, mli, na
Details for d3cmda1
PDB Entry: 3cmd (more details), 2.7 Å
PDB Description: Crystal structure of peptide deformylase from VRE-E.faecium
PDB Compounds: (A:) Peptide deformylaseSCOPe Domain Sequences for d3cmda1:
Sequence, based on SEQRES records: (download)
>d3cmda1 d.167.1.1 (A:1-187) automated matches {Enterococcus faecium [TaxId: 1352]}
mitmddiiregnptlrevakevslplseedislgkemleflknsqdpikaeelhlrggvg
laapqldiskriiavhvpspdpeadgpslstvmynpkilshsvqdaclgegegclsvdre
vpgyvvrhakitvsyydmngekhkirlknyesivvqheidhingvmfydhindqnpfalk
egvlvie
Sequence, based on observed residues (ATOM records): (download)
>d3cmda1 d.167.1.1 (A:1-187) automated matches {Enterococcus faecium [TaxId: 1352]}
mitmddiiregnptlrevakevslplseedislgkemleflknsqdpikaeelhlrggvg
laapqldiskriiavhvpsslstvmynpkilshsvqdaclgegegclsvdrevpgyvvrh
akitvsyydmngekhkirlknyesivvqheidhingvmfydhindqnpfalkegvlvie
SCOPe Domain Coordinates for d3cmda1:
Click to download the PDB-style file with coordinates for d3cmda1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3cmda1:
- d3cmda1 first appeared in SCOPe 2.03, called d3cmda_
- d3cmda1 appears in SCOPe 2.06
- d3cmda1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D