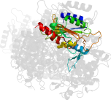
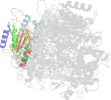
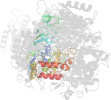
Lineage for d2zcid1 (2zci D:15-245)
- Root: SCOPe 2.04

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.109: PEP carboxykinase N-terminal domain [68922] (1 superfamily)
contains mixed beta-sheets; topology is partly similar to that of the catalytic C-terminal domain
Superfamily c.109.1: PEP carboxykinase N-terminal domain [68923] (2 families) 

Family c.109.1.0: automated matches [227144] (1 protein)
not a true family
Protein automated matches [226847] (4 species)
not a true protein
Species Corynebacterium glutamicum [TaxId:1718] [225441] (1 PDB entry) 
Domain d2zcid1: 2zci D:15-245 [207820]
Other proteins in same PDB: d2zcia2, d2zcib2, d2zcic2, d2zcid2
automated match to d1khba2
Details for d2zcid1
PDB Entry: 2zci (more details), 2.3 Å
PDB Description: Structure of a GTP-dependent bacterial PEP-carboxykinase from Corynebacterium glutamicum
PDB Compounds: (D:) Phosphoenolpyruvate carboxykinase [GTP]SCOPe Domain Sequences for d2zcid1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2zcid1 c.109.1.0 (D:15-245) automated matches {Corynebacterium glutamicum [TaxId: 1718]}
tknkellnwiadavelfqpeavvfvdgsqaewdrmaedlveagtliklneekrpnsylar
snpsdvarvesrtficsekeedagptnnwappqamkdemskhyagsmkgrtmyvvpfcmg
pisdpdpklgvqltdseyvvmsmrimtrmgiealdkigangsfvrclhsvgaplepgqed
vawpcndtkyitqfpetkeiwsygsgyggnailakkcyalriasvmareeg
SCOPe Domain Coordinates for d2zcid1:
Click to download the PDB-style file with coordinates for d2zcid1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2zcid1:
- d2zcid1 first appeared in SCOPe 2.03
- d2zcid1 appears in SCOPe 2.05
- d2zcid1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D