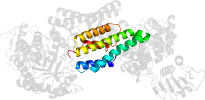
Lineage for d2vura3 (2vur A:496-624)
- Root: SCOPe 2.07

Class a: All alpha proteins [46456] (289 folds) 
Fold a.246: Hyaluronidase domain-like [140656] (3 superfamilies)
5 helices; bundle, closed, left-handed twist; up-and-down (meander) topology
Superfamily a.246.1: Hyaluronidase post-catalytic domain-like [140657] (2 families) 

Family a.246.1.1: Hyaluronidase post-catalytic domain-like [140658] (2 proteins) 
Protein Hyaluronidase, post-catalytic domain 3 [140659] (1 species) 
Species Clostridium perfringens [TaxId:1502] [140660] (7 PDB entries)
Uniprot Q8XL08 496-624
Domain d2vura3: 2vur A:496-624 [206490]
Other proteins in same PDB: d2vura1, d2vura2, d2vurb1, d2vurb2
automated match to d2cbia1
complexed with so4, yx1
Details for d2vura3
PDB Entry: 2vur (more details), 2.2 Å
PDB Description: chemical dissection of the link between streptozotocin, o-glcnac and pancreatic cell death
PDB Compounds: (A:) O-GlcNAcase nagJSCOPe Domain Sequences for d2vura3:
Sequence; same for both SEQRES and ATOM records: (download)
>d2vura3 a.246.1.1 (A:496-624) Hyaluronidase, post-catalytic domain 3 {Clostridium perfringens [TaxId: 1502]}
edapelrakmdelwnklsskedasalieelygefarmeeacnnlkanlpevaleecsrql
delitlaqgdkasldmivaqlnedteayesakeiaqnklntalssfavisekvaqsfiqe
alsfdltli
SCOPe Domain Coordinates for d2vura3:
Click to download the PDB-style file with coordinates for d2vura3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2vura3:
- d2vura3 first appeared in SCOPe 2.03
- d2vura3 appears in SCOPe 2.06
- d2vura3 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D