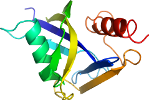Lineage for d2gb5a3 (2gb5 A:126-257)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.113: Nudix [55810] (1 superfamily)
beta(2)-alpha-beta(3)-alpha; 3 layers: alpha/beta/alpha; mixed sheet
contains beta-grasp motif
Superfamily d.113.1: Nudix [55811] (8 families) 

Family d.113.1.4: NADH pyrophosphatase [103214] (1 protein)
duplication: consists of two structurally similar domains separated by a rubredoxin-like zinc finger; the N-terminal domain has a rudiment Nudix fold, the C-terminal, probably catalytic, domain has the canonical fold
Protein NADH pyrophosphatase [103215] (1 species) 
Species Escherichia coli [TaxId:562] [103216] (2 PDB entries) 
Domain d2gb5a3: 2gb5 A:126-257 [204325]
Other proteins in same PDB: d2gb5a2, d2gb5a4, d2gb5b2, d2gb5b4
automated match to d1vk6a2
complexed with zn
Details for d2gb5a3
PDB Entry: 2gb5 (more details), 2.3 Å
PDB Description: Crystal structure of NADH pyrophosphatase (EC 3.6.1.22) (1790429) from Escherichia coli K12 at 2.30 A resolution
PDB Compounds: (A:) NADH pyrophosphataseSCOPe Domain Sequences for d2gb5a3:
Sequence; same for both SEQRES and ATOM records: (download)
>d2gb5a3 d.113.1.4 (A:126-257) NADH pyrophosphatase {Escherichia coli [TaxId: 562]}
qiapciivairrddsillaqhtrhrngvhtvlagfvevgetleqavarevmeesgikvkn
lryvtsqpwpfpqslmtafmaeydsgdividpkelleanwyryddlpllpppgtvarrli
edtvamcraeye
SCOPe Domain Coordinates for d2gb5a3:
Click to download the PDB-style file with coordinates for d2gb5a3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2gb5a3:
- d2gb5a3 first appeared in SCOPe 2.03
- d2gb5a3 appears in SCOPe 2.07
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d2gb5b1, d2gb5b2, d2gb5b3, d2gb5b4 |