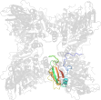
Lineage for d4hhhd2 (4hhh D:150-469)
- Root: SCOPe 2.04

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.1: TIM beta/alpha-barrel [51350] (33 superfamilies)
contains parallel beta-sheet barrel, closed; n=8, S=8; strand order 12345678
the first seven superfamilies have similar phosphate-binding sites
Superfamily c.1.14: RuBisCo, C-terminal domain [51649] (2 families) 
automatically mapped to Pfam PF00016
Family c.1.14.1: RuBisCo, large subunit, C-terminal domain [51650] (2 proteins)
N-terminal domain is alpha+beta- Protein automated matches [226984] (5 species)
not a true protein 
Species Pea (Pisum sativum) [TaxId:3888] [226514] (2 PDB entries) 
Domain d4hhhd2: 4hhh D:150-469 [202430]
Other proteins in same PDB: d4hhha1, d4hhhb1, d4hhhc1, d4hhhd1, d4hhhs_, d4hhht_, d4hhhu_, d4hhhv_
automated match to d1gk8a1
complexed with rub
Details for d4hhhd2
PDB Entry: 4hhh (more details), 2.2 Å
PDB Description: Structure of Pisum sativum Rubisco
PDB Compounds: (D:) ribulose bisphosphate carboxylase large chainSCOPe Domain Sequences for d4hhhd2:
Sequence; same for both SEQRES and ATOM records: (download)
>d4hhhd2 c.1.14.1 (D:150-469) automated matches {Pea (Pisum sativum) [TaxId: 3888]}
gpphgiqverdklnkygrpllgctikpklglsaknygravyeclrggldftkddenvnsq
pfmrwrdrflfcaeaiyksqaetgeikghylnatagtceemlkravfarelgvpivmhdy
ltggftanttlshycrdnglllhihramhavidrqknhgmhfrvlakalrlsggdhihag
tvvgklegereitlgfvdllrddyikkdrsrgiyftqdwvslpgvipvasggihvwhmpa
lteifgddsvlqfgggtlghpwgnapgavanrvaleacvqarnegrdlaregnaiireac
kwspelaaacevwkeikfef
SCOPe Domain Coordinates for d4hhhd2:
Click to download the PDB-style file with coordinates for d4hhhd2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4hhhd2:
- d4hhhd2 first appeared in SCOPe 2.03
- d4hhhd2 appears in SCOPe 2.05
- d4hhhd2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D