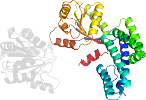Lineage for d1wp9e1 (1wp9 E:1-200)
- Root: SCOPe 2.03

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (25 families) 
division into families based on beta-sheet topologies
Family c.37.1.19: Tandem AAA-ATPase domain [81268] (24 proteins)
duplication: tandem repeat of two RecA-like (AAA) domains
Protein putative ATP-dependent RNA helicase PF2015 [142308] (1 species)
separate structures of other domains are known: the middle nuclease domain (89718) and the C-terminal RuvA-like HhH domain (PDB 1x2i)
Species Pyrococcus furiosus [TaxId:2261] [142309] (1 PDB entry)
Uniprot Q8TZH8 2-201! Uniprot Q8TZH8 202-487
Domain d1wp9e1: 1wp9 E:1-200 [197627]
automated match to d1wp9a1
complexed with po4
Details for d1wp9e1
PDB Entry: 1wp9 (more details), 2.9 Å
PDB Description: crystal structure of pyrococcus furiosus hef helicase domain
PDB Compounds: (E:) ATP-dependent RNA helicase, putativeSCOPe Domain Sequences for d1wp9e1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1wp9e1 c.37.1.19 (E:1-200) putative ATP-dependent RNA helicase PF2015 {Pyrococcus furiosus [TaxId: 2261]}
mvlrrdliqpriyqeviyakcketnclivlptglgktliammiaeyrltkyggkvlmlap
tkplvlqhaesfrrlfnlppekivaltgekspeerskawarakvivatpqtiendllagr
isledvslivfdeahravgnyayvfiareykrqaknplvigltaspgstpekimevinnl
giehieyrsenspdvrpyvk
SCOPe Domain Coordinates for d1wp9e1:
Click to download the PDB-style file with coordinates for d1wp9e1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1wp9e1:
- d1wp9e1 is new in SCOPe 2.03-stable
- d1wp9e1 appears in SCOPe 2.04
- d1wp9e1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D