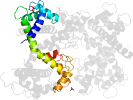
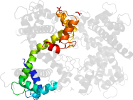
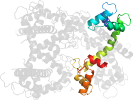Lineage for d4ds7d_ (4ds7 D:)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.39: EF Hand-like [47472] (4 superfamilies)
core: 4 helices; array of 2 hairpins, opened
Superfamily a.39.1: EF-hand [47473] (12 families) 
Duplication: consists of two EF-hand units: each is made of two helices connected with calcium-binding loop
Family a.39.1.5: Calmodulin-like [47502] (24 proteins)
Duplication: made with two pairs of EF-hands
Protein automated matches [190064] (21 species)
not a true protein
Species Kluyveromyces lactis [TaxId:284590] [195890] (1 PDB entry) 
Domain d4ds7d_: 4ds7 D: [195894]
automated match to d1lkja_
complexed with gol, so4, sr
Details for d4ds7d_
PDB Entry: 4ds7 (more details), 2.15 Å
PDB Description: crystal structure of yeast calmodulin bound to the c-terminal fragment of spindle pole body protein spc110
PDB Compounds: (D:) calmodulinSCOPe Domain Sequences for d4ds7d_:
Sequence; same for both SEQRES and ATOM records: (download)
>d4ds7d_ a.39.1.5 (D:) automated matches {Kluyveromyces lactis [TaxId: 284590]}
lteeqiaefkeafalfdkdnsgsisaselatvmrslglspseaevadlmneidvdgnhai
efseflalmsrqlkcndseqelleafkvfdkngdglisaaelkhvltsigekltdaevde
mlrevsdgsgeinikqfaallsk
SCOPe Domain Coordinates for d4ds7d_:
Click to download the PDB-style file with coordinates for d4ds7d_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4ds7d_:
- d4ds7d_ first appeared in SCOPe 2.02
- d4ds7d_ appears in SCOPe 2.07

 View in 3D
View in 3D