Lineage for d4esea_ (4ese A:)
- Root: SCOPe 2.02

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.23: Flavodoxin-like [52171] (15 superfamilies)
3 layers, a/b/a; parallel beta-sheet of 5 strand, order 21345
Superfamily c.23.5: Flavoproteins [52218] (9 families) 

Family c.23.5.3: Quinone reductase [52235] (4 proteins)
binds FAD
Protein automated matches [190235] (2 species)
not a true protein
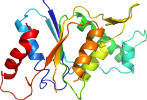
Species Yersinia pestis [TaxId:214092] [195328] (1 PDB entry) 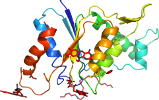
Domain d4esea_: 4ese A: [195329]
automated match to d1tika_
complexed with 12p, fmn, gol, imd
Details for d4esea_
PDB Entry: 4ese (more details), 1.45 Å
PDB Description: The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN.
PDB Compounds: (A:) FMN-dependent NADH-azoreductaseSCOPe Domain Sequences for d4esea_:
Sequence, based on SEQRES records: (download)
>d4esea_ c.23.5.3 (A:) automated matches {Yersinia pestis [TaxId: 214092]}
amskvlvlkssilatssqsnqladffveqwqaahagdqitvrdlaaqpipvldgelvgal
rpsgtaltprqqealalsdeliaelqandviviaapmynfniptqlknyfdmiaragvtf
rytekgpeglvtgkraiiltsrggihkdtptdlvvpylrlflgfigitdvefvfaegiay
gpevatkaqadaktllaqvva
Sequence, based on observed residues (ATOM records): (download)
>d4esea_ c.23.5.3 (A:) automated matches {Yersinia pestis [TaxId: 214092]}
amskvlvlkssilatssqsnqladffveqwqaahagdqitvrdlaaqpipvldgelvgal
rpsgtaltprqqealalsdeliaelqandviviaapmynfniptqlknyfdmiaragvtf
rytekgpeglvtgkraiiltsrgdlvvpylrlflgfigitdvefvfaegiaygpevatka
qadaktllaqvva
SCOPe Domain Coordinates for d4esea_:
Click to download the PDB-style file with coordinates for d4esea_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4esea_:
- d4esea_ is new in SCOPe 2.02-stable
- d4esea_ appears in SCOPe 2.03
- d4esea_ appears in the current release, SCOPe 2.08, called d4esea1
 View in 3D
View in 3D