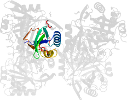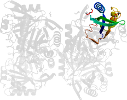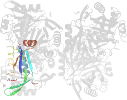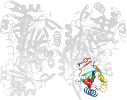Lineage for d3gsda_ (3gsd A:)
- Root: SCOPe 2.05

Class d: Alpha and beta proteins (a+b) [53931] (381 folds) 
Fold d.58: Ferredoxin-like [54861] (59 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.5: GlnB-like [54913] (6 families) 
form timeric structures with the orthogonally packed beta-sheets
Family d.58.5.2: Divalent ion tolerance proteins CutA (CutA1) [75434] (4 proteins) 
Protein automated matches [191031] (3 species)
not a true protein
Species Yersinia pestis [TaxId:214092] [188846] (1 PDB entry) 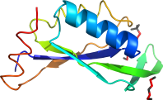
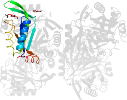
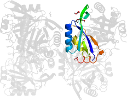
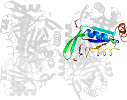
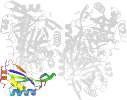
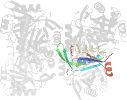
Domain d3gsda_: 3gsd A: [176950]
automated match to d1naqa_
complexed with edo, epe, na, peg
Details for d3gsda_
PDB Entry: 3gsd (more details), 2.05 Å
PDB Description: 2.05 angstrom structure of a divalent-cation tolerance protein (cuta) from yersinia pestis
PDB Compounds: (A:) Divalent-cation tolerance protein cutASCOPe Domain Sequences for d3gsda_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3gsda_ d.58.5.2 (A:) automated matches {Yersinia pestis [TaxId: 214092]}
ysnaivvlctapdeasaqnlaaqvlgeklaacvtllpgatslyywegkleqeyevqllfk
sntdhqqalltyikqhhpyqtpellvlpvrdgdkdylswlnasll
SCOPe Domain Coordinates for d3gsda_:
Click to download the PDB-style file with coordinates for d3gsda_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3gsda_:
- d3gsda_ first appeared in SCOPe 2.01
- d3gsda_ appears in SCOPe 2.04
- d3gsda_ appears in SCOPe 2.06
- d3gsda_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D