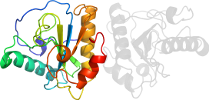
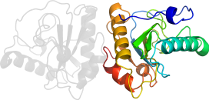
Lineage for d2eava_ (2eav A:)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.118: N-acetylmuramoyl-L-alanine amidase-like [55845] (1 superfamily)
contains mixed beta-sheet
Superfamily d.118.1: N-acetylmuramoyl-L-alanine amidase-like [55846] (2 families) 

Family d.118.1.1: N-acetylmuramoyl-L-alanine amidase-like [55847] (11 proteins)
Family 2 zinc amidase;
Protein automated matches [190549] (4 species)
not a true protein
Species Human (Homo sapiens) [TaxId:9606] [187654] (1 PDB entry) 
Domain d2eava_: 2eav A: [163925]
automated match to d1sk3a_
complexed with ni
Details for d2eava_
PDB Entry: 2eav (more details), 2.2 Å
PDB Description: Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ibeta
PDB Compounds: (A:) Peptidoglycan recognition protein-I-betaSCOPe Domain Sequences for d2eava_:
Sequence; same for both SEQRES and ATOM records: (download)
>d2eava_ d.118.1.1 (A:) automated matches {Human (Homo sapiens) [TaxId: 9606]}
cpgivprsvwgarethcprmtlpakygiiihtagrtcnisdecrllvrdiqsfyidrlks
cdigynflvgqdgaiyegvgwnvqgsstpgyddialgitfmgtftgippnaaaleaaqdl
iqcamvkgyltpnyllvghsdvartlspgqalyniistwphfkh
SCOPe Domain Coordinates for d2eava_:
Click to download the PDB-style file with coordinates for d2eava_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2eava_:
- d2eava_ first appeared in SCOPe 2.01
- d2eava_ appears in SCOPe 2.07
 View in 3D
View in 3D