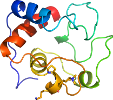
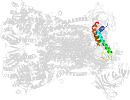
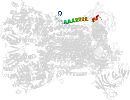
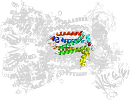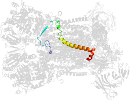Lineage for d3cxhw_ (3cxh W:)
- Root: SCOPe 2.07

Class a: All alpha proteins [46456] (289 folds) 
Fold a.3: Cytochrome c [46625] (1 superfamily)
core: 3 helices; folded leaf, opened
Superfamily a.3.1: Cytochrome c [46626] (9 families) 
covalently-bound heme completes the core
Family a.3.1.1: monodomain cytochrome c [46627] (16 proteins) 
Protein Mitochondrial cytochrome c [46642] (7 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [46643] (90 PDB entries)
Uniprot P00044
Domain d3cxhw_: 3cxh W: [161631]
Other proteins in same PDB: d3cxha1, d3cxha2, d3cxhb1, d3cxhb2, d3cxhc1, d3cxhc2, d3cxhd1, d3cxhd2, d3cxhe1, d3cxhe2, d3cxhf_, d3cxhg_, d3cxhh_, d3cxhi_, d3cxhj_, d3cxhk_, d3cxhl1, d3cxhl2, d3cxhm1, d3cxhm2, d3cxhn1, d3cxhn2, d3cxho1, d3cxho2, d3cxhp1, d3cxhp2, d3cxhq_, d3cxhr_, d3cxhs_, d3cxht_, d3cxhu_, d3cxhv_
automated match to d1yeaa_
complexed with 6ph, 7ph, 8pe, 9pe, cn6, fes, hem, sma, suc, umq
Details for d3cxhw_
PDB Entry: 3cxh (more details), 2.5 Å
PDB Description: structure of yeast complex iii with isoform-2 cytochrome c bound and definition of a minimal core interface for electron transfer.
PDB Compounds: (W:) Cytochrome c iso-2SCOPe Domain Sequences for d3cxhw_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3cxhw_ a.3.1.1 (W:) Mitochondrial cytochrome c {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
akestgfkpgsakkgatlfktrcqqchtieeggpnkvgpnlhgifgrhsgqvkgysytda
ninknvkwdedsmseyltnpkkyipgtkmafaglkkekdrndlitymtkaak
SCOPe Domain Coordinates for d3cxhw_:
Click to download the PDB-style file with coordinates for d3cxhw_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3cxhw_:
- d3cxhw_ first appeared in SCOPe 2.01
- d3cxhw_ appears in SCOPe 2.06
- d3cxhw_ appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d3cxha1, d3cxha2, d3cxhb1, d3cxhb2, d3cxhc1, d3cxhc2, d3cxhd1, d3cxhd2, d3cxhe1, d3cxhe2, d3cxhf_, d3cxhg_, d3cxhh_, d3cxhi_, d3cxhj_, d3cxhk_, d3cxhl1, d3cxhl2, d3cxhm1, d3cxhm2, d3cxhn1, d3cxhn2, d3cxho1, d3cxho2, d3cxhp1, d3cxhp2, d3cxhq_, d3cxhr_, d3cxhs_, d3cxht_, d3cxhu_, d3cxhv_ |