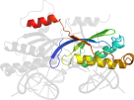
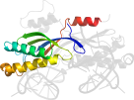
Lineage for d2viha1 (2vih A:6-131)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.58: Ferredoxin-like [54861] (59 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.57: Transposase IS200-like [143422] (1 family) 
contains extra N-terminal hairpin and C-terminal helix, both are involved in dimerization; there can be helix-swapping in the dimer
Family d.58.57.1: Transposase IS200-like [143423] (3 proteins)
Pfam PF01797
Protein ISHP608 transposase [143426] (1 species) 
Species Helicobacter pylori [TaxId:210] [143427] (7 PDB entries)
Uniprot Q933Z0 4-133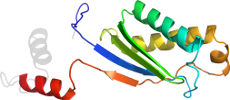
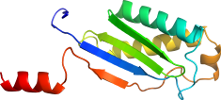
Domain d2viha1: 2vih A:6-131 [153168]
automatically matched to d2a6ma1
Details for d2viha1
PDB Entry: 2vih (more details), 2.1 Å
PDB Description: crystal structure of the is608 transposase in complex with left end 26-mer dna
PDB Compounds: (A:) transposase orfaSCOP Domain Sequences for d2viha1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2viha1 d.58.57.1 (A:6-131) ISHP608 transposase {Helicobacter pylori [TaxId: 210]}
lyksnhnvvysckyhivwcpkyrrkvlvgavemrlkeiiqevakelrveiiemqtdkdhi
hiladidpsfgvmkfiktakgrssrilrqefnhlktklptlwtnscfistvggaplnvvk
qyienq
SCOP Domain Coordinates for d2viha1:
Click to download the PDB-style file with coordinates for d2viha1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2viha1:
- d2viha1 is new in SCOP 1.75
- d2viha1 appears in SCOPe 2.01
- d2viha1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D