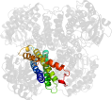
Lineage for d2reha1 (2reh A:261-431)
- Root: SCOPe 2.06

Class a: All alpha proteins [46456] (289 folds) 
Fold a.29: Bromodomain-like [47363] (15 superfamilies)
4 helices; bundle; minor mirror variant of up-and-down topology
Superfamily a.29.3: Acyl-CoA dehydrogenase C-terminal domain-like [47203] (3 families) 
multidomain flavoprotein; N-terminal domain is all-alpha; the middle domain is open (5,8) barrel
Family a.29.3.1: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain [47204] (10 proteins) 
Protein automated matches [231931] (2 species)
not a true protein
Species Fungus (Fusarium oxysporum) [TaxId:5507] [231932] (6 PDB entries) 
Domain d2reha1: 2reh A:261-431 [151973]
Other proteins in same PDB: d2reha2, d2rehb2, d2rehc2, d2rehd2
automated match to d2c0ua1
complexed with fad
Details for d2reha1
PDB Entry: 2reh (more details), 2.4 Å
PDB Description: mechanistic and structural analyses of the roles of arg409 and asp402 in the reaction of the flavoprotein nitroalkane oxidase
PDB Compounds: (A:) nitroalkane oxidaseSCOPe Domain Sequences for d2reha1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2reha1 a.29.3.1 (A:261-431) automated matches {Fungus (Fusarium oxysporum) [TaxId: 5507]}
pglkaqglvetafamsaalvgamaigtaraafeealvfaksdtrggskhiiehqsvadkl
idckirletsrllvwkavttledealewkvklemamqtkiyttdvavecvidamkavgmk
syakdmsfprllnevmcyplfeggniglrrrqmqrvmaledyepwaatygs
SCOPe Domain Coordinates for d2reha1:
Click to download the PDB-style file with coordinates for d2reha1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2reha1:
- d2reha1 first appeared in SCOP 1.75
- d2reha1 appears in SCOPe 2.05
- d2reha1 appears in SCOPe 2.07
- d2reha1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D