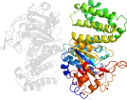Lineage for d2radb1 (2rad B:40-442)
- Root: SCOP 1.75

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.150: EreA/ChaN-like [159500] (1 superfamily)
Core: 3 layers: a/b/a; parallel beta-sheet of 5 strands, order:51423;
Superfamily c.150.1: EreA/ChaN-like [159501] (3 families) 
there are four conserved residues in the putative active site: two His and two Glu
Family c.150.1.3: EreA-like [159508] (2 proteins)
Pfam PF05139; Erythromycin esterase-like; the superfamily core is decorated with insertion of a four-helical bundle and a C-terminal alpha+beta extension
Protein Succinoglycan biosynthesis protein BC3120 [159509] (1 species) 
Species Bacillus cereus [TaxId:1396] [159510] (2 PDB entries)
Uniprot Q81BN2 40-442
Domain d2radb1: 2rad B:40-442 [151822]
automatically matched to 2RAD A:40-442
Details for d2radb1
PDB Entry: 2rad (more details), 2.75 Å
PDB Description: Crystal structure of the succinoglycan biosynthesis protein. Northeast structural Genomics Consortium target BcR135
PDB Compounds: (B:) Succinoglycan biosynthesis proteinSCOP Domain Sequences for d2radb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2radb1 c.150.1.3 (B:40-442) Succinoglycan biosynthesis protein BC3120 {Bacillus cereus [TaxId: 1396]}
nqiakwleahakplkttnptaslndlkplknmvgsasivglgeathgahevftmkhrivk
ylvsekgftnlvleegwdraleldryvltgkgnpsqhltpvfktkemldlldwirqynan
pkhkskvrvigmdiqsvnenvynniieyikannskllprveekikglipvtkdmntfesl
tkeekekyvldaktisalleenksylngkskefawikqnariieqfttmlatppdkpadf
ylkhdiamyenakwteehlgktivwghnghvsktnmlsfiypkvagqhlaeyygkryvsi
gtsvyegqynvknsdgefgpygtlksddpnsynyifgqvkkdqffidlrkangvtktwln
eqhpifagittegpdipktvdislgkafdilvqiqkvspsqvh
SCOP Domain Coordinates for d2radb1:
Click to download the PDB-style file with coordinates for d2radb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2radb1:
- d2radb1 is new in SCOP 1.75
- d2radb1 appears in SCOPe 2.01
- d2radb1 appears in the current release, SCOPe 2.08, called d2radb_
 View in 3D
View in 3D