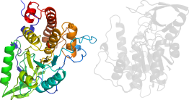Lineage for d2pt0b_ (2pt0 B:)
- Root: SCOPe 2.06

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.45: (Phosphotyrosine protein) phosphatases II [52798] (1 superfamily)
core: 3 layers, a/b/a; parallel beta-sheet of 4 strands, order 1423
Superfamily c.45.1: (Phosphotyrosine protein) phosphatases II [52799] (6 families) 
share with the family I the common active site structure with a circularly permuted topology
Family c.45.1.4: Myo-inositol hexaphosphate phosphohydrolase (phytase) PhyA [117583] (2 proteins) 
Protein Myo-inositol hexaphosphate phosphohydrolase (phytase) PhyA [117584] (1 species) 
Species Selenomonas ruminantium [TaxId:971] [117585] (12 PDB entries)
Uniprot Q7WUJ1 34-346
Domain d2pt0b_: 2pt0 B: [149833]
automated match to d3moza_
complexed with gol
Details for d2pt0b_
PDB Entry: 2pt0 (more details), 1.7 Å
PDB Description: Structure of Selenomonas ruminantium PTP-like phytase with the active site cysteine oxidized to cysteine-sulfonic acid
PDB Compounds: (B:) myo-inositol hexaphosphate phosphohydrolaseSCOPe Domain Sequences for d2pt0b_:
Sequence; same for both SEQRES and ATOM records: (download)
>d2pt0b_ c.45.1.4 (B:) Myo-inositol hexaphosphate phosphohydrolase (phytase) PhyA {Selenomonas ruminantium [TaxId: 971]}
tvtepvgsyaraerpqdfegfvwrldndgkealprnfrtsadalrapekkfhldaayvps
regmdalhisgssaftpaqlknvaaklrektagpiydvdlrqeshgyldgipvswygerd
wanlgksqhealaderhrlhaalhktvyiaplgkhklpeggevrrvqkvqteqevaeaag
mryfriaatdhvwptpenidrflafyrtlpqdawlhfhceagvgrttafmvmtdmlknps
vslkdilyrqheiggfyygefpiktkdkdswktkyyrekivmieqfyryvqenradgyqt
pwsvwlkshpaka
SCOPe Domain Coordinates for d2pt0b_:
Click to download the PDB-style file with coordinates for d2pt0b_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2pt0b_:
- d2pt0b_ first appeared in SCOP 1.75, called d2pt0b1
- d2pt0b_ appears in SCOPe 2.05
- d2pt0b_ appears in SCOPe 2.07
- d2pt0b_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D