Lineage for d2pkhd1 (2pkh D:109-249)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.190: Chorismate lyase-like [64287] (1 superfamily)
duplication of alpha(2)-beta(3) motif; antiparallel beta sheet, order 123654
Superfamily d.190.1: Chorismate lyase-like [64288] (3 families) 

Family d.190.1.2: UTRA domain [143473] (10 proteins)
Pfam PF07702
Protein Histidine utilization repressor HutC [160399] (1 species) 
Species Pseudomonas syringae pv. tomato [TaxId:323] [160400] (1 PDB entry)
Uniprot Q87UX0 109-249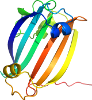
Domain d2pkhd1: 2pkh D:109-249 [149599]
automatically matched to 2PKH A:109-249
complexed with edo
Details for d2pkhd1
PDB Entry: 2pkh (more details), 1.95 Å
PDB Description: Structural Genomics, the crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000
PDB Compounds: (D:) Histidine utilization repressorSCOP Domain Sequences for d2pkhd1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2pkhd1 d.190.1.2 (D:109-249) Histidine utilization repressor HutC {Pseudomonas syringae pv. tomato [TaxId: 323]}
rhtckvmvlkeeaagseralaldmregqrvfhslivhfendipvqiedrfvnaqvapdyl
kqdftlqtpyaylsqvapltegehvveailaeadeckllqidagepcllirrrtwsgrqp
vtaarlihpgsrhrlegrftk
SCOP Domain Coordinates for d2pkhd1:
Click to download the PDB-style file with coordinates for d2pkhd1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2pkhd1:
- d2pkhd1 is new in SCOP 1.75
- d2pkhd1 is called d2pkhd_ in SCOPe 2.01
- d2pkhd1 appears in the current release, SCOPe 2.08, called d2pkhd_
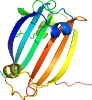
 View in 3D
View in 3D