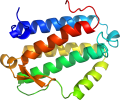
Lineage for d2nsga1 (2nsg A:1-160)
- Root: SCOPe 2.01

Class a: All alpha proteins [46456] (284 folds) 
Fold a.213: DinB/YfiT-like putative metalloenzymes [109853] (1 superfamily)
core: 4 helices; bundle, closed, left-handed twist; an unusual topology with a higher contact order
Superfamily a.213.1: DinB/YfiT-like putative metalloenzymes [109854] (4 families) 
contains metal-binding site on the bundle surface surrounded by loops
Family a.213.1.4: Maleylpyruvate isomerase-like [158522] (1 protein) 
Protein Micothiol-dependent maleylpyruvate isomerase [158523] (1 species) 
Species Corynebacterium glutamicum [TaxId:1718] [158524] (2 PDB entries)
Uniprot Q8NLC1 1-160
Cgl3021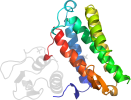
Domain d2nsga1: 2nsg A:1-160 [148388]
Other proteins in same PDB: d2nsga2
complexed with gol, so4; mutant
Details for d2nsga1
PDB Entry: 2nsg (more details), 2.05 Å
PDB Description: crystal structure of the mycothiol-dependent maleylpyruvate isomerase h52a mutant
PDB Compounds: (A:) Hypothetical protein Cgl3021SCOPe Domain Sequences for d2nsga1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2nsga1 a.213.1.4 (A:1-160) Micothiol-dependent maleylpyruvate isomerase {Corynebacterium glutamicum [TaxId: 1718]}
mttfhdlpleerltlarlgtshysrqlslvdnaefgehsllegwtrshliaavaynaial
cnlmhwantgeetpmyvspearneeiaygstlnpdalrnlhehsvarldvawretsedaw
shevltaqgrtvpasetlwmrsrevwihavdlgavatfgd
SCOPe Domain Coordinates for d2nsga1:
Click to download the PDB-style file with coordinates for d2nsga1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2nsga1:
- d2nsga1 first appeared in SCOP 1.75
- d2nsga1 appears in SCOPe 2.02
- d2nsga1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D