Lineage for d2ecoa1 (2eco A:1-302)
- Root: SCOP 1.75

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.79: Tryptophan synthase beta subunit-like PLP-dependent enzymes [53685] (1 superfamily)
consists of two similar domains related by pseudo dyad; duplication
core: 3 layers, a/b/a; parallel beta-sheet of 4 strands, order 3214
Superfamily c.79.1: Tryptophan synthase beta subunit-like PLP-dependent enzymes [53686] (1 family) 

Family c.79.1.1: Tryptophan synthase beta subunit-like PLP-dependent enzymes [53687] (8 proteins) 
Protein O-acetylserine sulfhydrylase (Cysteine synthase) [53690] (7 species) 
Species Thermus thermophilus [TaxId:274] [142743] (4 PDB entries)
Uniprot Q5SLE6 1-302
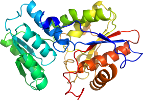
Domain d2ecoa1: 2eco A:1-302 [146790]
automatically matched to d1ve1a1
complexed with 4mv, plp
Details for d2ecoa1
PDB Entry: 2eco (more details), 1.9 Å
PDB Description: Crystal Structure of T.th. HB8 O-acetylserine sulfhydrylase Complexed with 4-methylvalerate
PDB Compounds: (A:) O-acetylserine (Thiol)-lyaseSCOP Domain Sequences for d2ecoa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2ecoa1 c.79.1.1 (A:1-302) O-acetylserine sulfhydrylase (Cysteine synthase) {Thermus thermophilus [TaxId: 274]}
mrvegaigktpvvrlakvvepdmaevwvkleglnpggsikdrpawymikdaeergilrpg
sgqviveptsgntgiglamiaasrgyrliltmpaqmseerkrvlkafgaelvltdperrm
laareealrlkeelgafmpdqfknpanvrahyettgpelyealegridafvygsgtggti
tgvgrylkeriphvkviaveparsnvlsggkmgqhgfqgmgpgfipenldlslldgviqv
weedafplarrlareeglflgmssggivwaalqvarelgpgkrvacispdggwkylstpl
ya
SCOP Domain Coordinates for d2ecoa1:
Click to download the PDB-style file with coordinates for d2ecoa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2ecoa1:
- d2ecoa1 is new in SCOP 1.75
- d2ecoa1 is called d2ecoa_ in SCOPe 2.01
- d2ecoa1 appears in the current release, SCOPe 2.08, called d2ecoa_
 View in 3D
View in 3D