Lineage for d2e1ba2 (2e1b A:88-216)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.67: RRF/tRNA synthetase additional domain-like [55185] (4 superfamilies)
core: alpha-beta(2)-alpha-beta(2); 2 layers: alpha/beta
Superfamily d.67.1: ThrRS/AlaRS common domain [55186] (2 families) 
putative editing domain found in the N-terminal part of ThrRS, the C-terminal of AlaRS, and as a stand-alone protein; probable circular permutation of LuxS (d.185.1.2)
Family d.67.1.2: AlaX-like [103051] (2 proteins) 
Protein AlaX-M trans-editing enzyme, C-terminal domain [160440] (1 species) 
Species Pyrococcus horikoshii [TaxId:53953] [160441] (1 PDB entry)
Uniprot O57848 88-216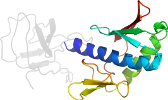
Domain d2e1ba2: 2e1b A:88-216 [146624]
Other proteins in same PDB: d2e1ba1
complexed with zn
Details for d2e1ba2
PDB Entry: 2e1b (more details), 2.7 Å
PDB Description: Crystal structure of the AlaX-M trans-editing enzyme from Pyrococcus horikoshii
PDB Compounds: (A:) 216aa long hypothetical alanyl-tRNA synthetaseSCOP Domain Sequences for d2e1ba2:
Sequence; same for both SEQRES and ATOM records: (download)
>d2e1ba2 d.67.1.2 (A:88-216) AlaX-M trans-editing enzyme, C-terminal domain {Pyrococcus horikoshii [TaxId: 53953]}
dwdyryklmrihtglhllehvlnevlgegnwqlvgsgmsvekgrydiaypenlnkykeqi
islfnkyvdeggevkiwwegdrrytqirdfevipcggthvkdikeighikklkrssigrg
kqrlemwle
SCOP Domain Coordinates for d2e1ba2:
Click to download the PDB-style file with coordinates for d2e1ba2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2e1ba2:
- d2e1ba2 is new in SCOP 1.75
- d2e1ba2 appears in SCOPe 2.01
- d2e1ba2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D