Lineage for Protein: Erythromycin synthase, eryAI, 1st ketoreductase module
- Root: SCOPe 2.01

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.2: NAD(P)-binding Rossmann-fold domains [51734] (1 superfamily)
core: 3 layers, a/b/a; parallel beta-sheet of 6 strands, order 321456
The nucleotide-binding modes of this and the next two folds/superfamilies are similar
Superfamily c.2.1: NAD(P)-binding Rossmann-fold domains [51735] (13 families) 
Family c.2.1.2: Tyrosine-dependent oxidoreductases [51751] (71 proteins)
also known as short-chain dehydrogenases and SDR family
parallel beta-sheet is extended by 7th strand, order 3214567; left-handed crossover connection between strands 6 and 7
Protein Erythromycin synthase, eryAI, 1st ketoreductase module [141909] (1 species)
tandem repeat of two SDR-like modules resembling a dimer commonly founf in the family; only the second module retains the coenzyme-binding site
Species:

Saccharopolyspora erythraea [TaxId:1836] [141910] (2 PDB entries)
Uniprot Q5UNP6 1448-1656! Uniprot Q5UNP6 1657-1915- Domains for 2fr0:
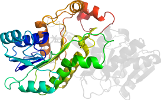
Domain d2fr0a1: 2fr0 A:1657-1915 [133959]
complexed with ndp
Domain d2fr0a2: 2fr0 A:1448-1656 [133960]
complexed with ndp
- Domains for 2fr1:
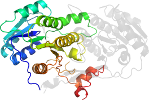
Domain d2fr1a1: 2fr1 A:1657-1915 [133961]
automatically matched to 2FR0 A:1657-1915
complexed with ndp
Domain d2fr1a2: 2fr1 A:1448-1656 [133962]
automatically matched to 2FR0 A:1448-1656
complexed with ndp
- Domains for 2fr0:
More info for Protein Erythromycin synthase, eryAI, 1st ketoreductase module from c.2.1.2: Tyrosine-dependent oxidoreductases
Timeline for Protein Erythromycin synthase, eryAI, 1st ketoreductase module from c.2.1.2: Tyrosine-dependent oxidoreductases:
- Protein Erythromycin synthase, eryAI, 1st ketoreductase module from c.2.1.2: Tyrosine-dependent oxidoreductases first appeared in SCOP 1.73
- Protein Erythromycin synthase, eryAI, 1st ketoreductase module from c.2.1.2: Tyrosine-dependent oxidoreductases appears in SCOP 1.75
- Protein Erythromycin synthase, eryAI, 1st ketoreductase module from c.2.1.2: Tyrosine-dependent oxidoreductases appears in SCOPe 2.02
- Protein Erythromycin synthase, eryAI, 1st ketoreductase module from c.2.1.2: Tyrosine-dependent oxidoreductases last appears in SCOPe 2.07