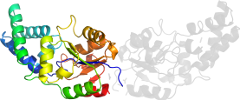
Lineage for d2hszb1 (2hsz B:1-224)
- Root: SCOP 1.73

Class c: Alpha and beta proteins (a/b) [51349] (141 folds) 
Fold c.108: HAD-like [56783] (1 superfamily)
3 layers: a/b/a; parallel beta-sheet of 6 strands, order 321456
Superfamily c.108.1: HAD-like [56784] (23 families) 
usually contains an insertion (sub)domain after strand 1
Family c.108.1.6: beta-Phosphoglucomutase-like [75173] (9 proteins)
the insertion subdomain is a 4-helical bundle
Protein Phosphoglycolate phosphatase Gph [142143] (1 species) 
Species Haemophilus somnus [TaxId:731] [142144] (1 PDB entry) 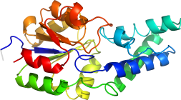
Domain d2hszb1: 2hsz B:1-224 [136727]
automatically matched to 2HSZ A:1-224
complexed with act, cl, unl
Details for d2hszb1
PDB Entry: 2hsz (more details), 1.9 Å
PDB Description: crystal structure of a predicted phosphoglycolate phosphatase (hs_0176) from haemophilus somnus 129pt at 1.90 a resolution
PDB Compounds: (B:) novel predicted phosphataseSCOP Domain Sequences for d2hszb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2hszb1 c.108.1.6 (B:1-224) Phosphoglycolate phosphatase Gph {Haemophilus somnus [TaxId: 731]}
mtqfkligfdldgtlvnslpdlalsinsalkdvnlpqasenlvmtwigngadvlsqravd
wackqaekeltedefkyfkrqfgfyygenlcnisrlypnvketlealkaqgyilavvtnk
ptkhvqpiltafgidhlfsemlggqslpeikphpapfyylcgkfglypkqilfvgdsqnd
ifaahsagcavvgltygynynipiaqskpdwifddfadilkitq
SCOP Domain Coordinates for d2hszb1:
Click to download the PDB-style file with coordinates for d2hszb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2hszb1:
- d2hszb1 is new in SCOP 1.73
- d2hszb1 appears in SCOP 1.75
- d2hszb1 appears in the current release, SCOPe 2.08, called d2hszb2

 View in 3D
View in 3D