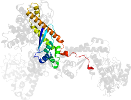
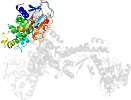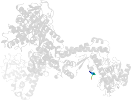Lineage for d2h23b1 (2h23 B:311-482)
- Root: SCOPe 2.06

Class a: All alpha proteins [46456] (289 folds) 
Fold a.166: RuBisCo LSMT C-terminal, substrate-binding domain [81821] (1 superfamily)
multihelical; irregular array of long and short helices
Superfamily a.166.1: RuBisCo LSMT C-terminal, substrate-binding domain [81822] (1 family) 

Family a.166.1.1: RuBisCo LSMT C-terminal, substrate-binding domain [81823] (1 protein) 
Protein RuBisCo LSMT C-terminal, substrate-binding domain [81824] (1 species) 
Species Pea (Pisum sativum) [TaxId:3888] [81825] (7 PDB entries) 
Domain d2h23b1: 2h23 B:311-482 [135984]
Other proteins in same PDB: d2h23a2, d2h23a3, d2h23b2, d2h23b3, d2h23c2, d2h23c3
automated match to d1p0yb1
complexed with m3l, sah
Details for d2h23b1
PDB Entry: 2h23 (more details), 2.45 Å
PDB Description: structure of rubisco lsmt bound to trimethyllysine and adohcy
PDB Compounds: (B:) Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferaseSCOPe Domain Sequences for d2h23b1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2h23b1 a.166.1.1 (B:311-482) RuBisCo LSMT C-terminal, substrate-binding domain {Pea (Pisum sativum) [TaxId: 3888]}
aytltleisesdpffddkldvaesngfaqtayfdifynrtlppgllpylrlvalggtdaf
lleslfrdtiwghlelsvsrdneellckavreacksalagyhttieqdrelkegnldsrl
aiavgiregekmvlqqidgifeqkeleldqleyyqerrlkdlglcgengdil
SCOPe Domain Coordinates for d2h23b1:
Click to download the PDB-style file with coordinates for d2h23b1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2h23b1:
- d2h23b1 first appeared in SCOP 1.73
- d2h23b1 appears in SCOPe 2.05
- d2h23b1 appears in SCOPe 2.07
- d2h23b1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D