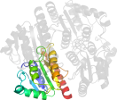
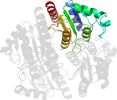
Lineage for d2gswa1 (2gsw A:3-170)
- Root: SCOP 1.73

Class c: Alpha and beta proteins (a/b) [51349] (141 folds) 
Fold c.23: Flavodoxin-like [52171] (15 superfamilies)
3 layers, a/b/a; parallel beta-sheet of 5 strand, order 21345
Superfamily c.23.5: Flavoproteins [52218] (8 families) 

Family c.23.5.4: NADPH-dependent FMN reductase [89590] (4 proteins)
Pfam PF03358
Protein Azobenzene reductase [89591] (1 species) 
Species Bacillus subtilis [TaxId:1423] [89592] (2 PDB entries)
YhdA gene product
Domain d2gswa1: 2gsw A:3-170 [135595]
automatically matched to d1nni1_
complexed with fmn
Details for d2gswa1
PDB Entry: 2gsw (more details), 2.92 Å
PDB Description: Crystal Structure of the Putative NADPH-dependent Azobenzene FMN-Reductase YhdA from Bacillus subtilis, Northeast Structural Genomics Target SR135
PDB Compounds: (A:) yhdASCOP Domain Sequences for d2gswa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2gswa1 c.23.5.4 (A:3-170) Azobenzene reductase {Bacillus subtilis [TaxId: 1423]}
mlvingtprkhgrtriaasyiaalyhtdlidlsefvlpvfngeaeqsellkvqelkqrvt
kadaivllspeyhsgmsgalknaldflsseqfkykpvallavagggkgginalnnmrtvm
rgvyanvipkqlvldpvhidvenatvaenikesikelveelsmfakag
SCOP Domain Coordinates for d2gswa1:
Click to download the PDB-style file with coordinates for d2gswa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2gswa1:
- d2gswa1 is new in SCOP 1.73
- d2gswa1 appears in SCOP 1.75
- d2gswa1 appears in the current release, SCOPe 2.08, called d2gswa_

 View in 3D
View in 3D