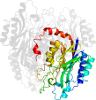
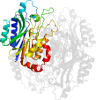
Lineage for d2ggla1 (2ggl A:3-304)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.160: Carbon-nitrogen hydrolase [56316] (1 superfamily)
4 layers: alpha/beta/beta/alpha; mixed beta sheets; contains duplication
Superfamily d.160.1: Carbon-nitrogen hydrolase [56317] (3 families) 
Pfam PF00795; some topological similarities to the metallo-dependent phosphatases and DNase I-like nucleases
Family d.160.1.2: Carbamilase [64433] (3 proteins) 
Protein N-carbamoyl-D-aminoacid amidohydrolase [64434] (2 species) 
Species Agrobacterium radiobacter [TaxId:358] [64436] (7 PDB entries) 
Domain d2ggla1: 2ggl A:3-304 [135148]
mutant
Details for d2ggla1
PDB Entry: 2ggl (more details), 2.4 Å
PDB Description: the mutant a222c of agrobacterium radiobacter n-carbamoyl-d-amino acid amidohydrolase
PDB Compounds: (A:) N-carbamoyl-D-amino acid amidohydrolaseSCOPe Domain Sequences for d2ggla1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2ggla1 d.160.1.2 (A:3-304) N-carbamoyl-D-aminoacid amidohydrolase {Agrobacterium radiobacter [TaxId: 358]}
rqmilavgqqgpiaraetreqvvgrlldmltnaasrgvnfivfpelalttffprwhftde
aeldsfyetempgpvvrplfetaaelgigfnlgyaelvveggvkrrfntsilvdksgkiv
gkyrkihlpghkeyeayrpfqhlekryfepgdlgfpvydvdaakmgmficndrrwpetwr
vmglkgaeiicggyntpthnppvpqhdhltsfhhllsmqcgsyqngawsaaagkvgmeeg
cmllghscivaptgeivaltttledevitaaldldrcrelrehifnfkahrqpqhyglia
ef
SCOPe Domain Coordinates for d2ggla1:
Click to download the PDB-style file with coordinates for d2ggla1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2ggla1:
- d2ggla1 first appeared in SCOP 1.73
- d2ggla1 appears in SCOPe 2.06
- d2ggla1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D