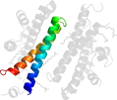
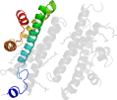
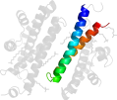
Lineage for d2f6ma2 (2f6m A:322-385)
- Root: SCOPe 2.06

Class a: All alpha proteins [46456] (289 folds) 
Fold a.2: Long alpha-hairpin [46556] (20 superfamilies)
2 helices; antiparallel hairpin, left-handed twist
Superfamily a.2.17: Endosomal sorting complex assembly domain [140111] (3 families) 
forms 'fan'-like heterooligomers, in which the domains of different subunit make similar interactions at the tip of the hairpin
Family a.2.17.1: VPS23 C-terminal domain [140112] (1 protein)
automatically mapped to Pfam PF09454
Protein Vacuolar protein sorting-associated protein 23, VPS23 [140113] (1 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [140114] (3 PDB entries)
Uniprot P25604 322-385! Uniprot P25604 325-383
Domain d2f6ma2: 2f6m A:322-385 [133051]
Other proteins in same PDB: d2f6ma3, d2f6mb2, d2f6mb3, d2f6mc3, d2f6md_
automated match to d2f66a1
complexed with ddq, mg
Details for d2f6ma2
PDB Entry: 2f6m (more details), 2.1 Å
PDB Description: structure of a vps23-c:vps28-n subcomplex
PDB Compounds: (A:) suppressor protein stp22 of temperature-sensitive alpha-factor receptor and arginine permeaseSCOPe Domain Sequences for d2f6ma2:
Sequence; same for both SEQRES and ATOM records: (download)
>d2f6ma2 a.2.17.1 (A:322-385) Vacuolar protein sorting-associated protein 23, VPS23 {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
tdglnqlynlvaqdyaltdtiealsrmlhrgtipldtfvkqgrelarqqflvrwhiqrit
spls
SCOPe Domain Coordinates for d2f6ma2:
Click to download the PDB-style file with coordinates for d2f6ma2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2f6ma2:
- d2f6ma2 first appeared in SCOP 1.73, called d2f6ma1
- d2f6ma2 was called d2f6ma_ in SCOPe 2.05
- d2f6ma2 appears in SCOPe 2.07
- d2f6ma2 appears in the current release, SCOPe 2.08
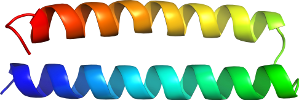
 View in 3D
View in 3D