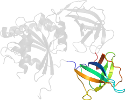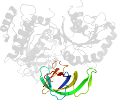Lineage for d1zunb2 (1zun B:330-434)
- Root: SCOP 1.73

Class b: All beta proteins [48724] (165 folds) 
Fold b.44: Elongation factor/aminomethyltransferase common domain [50464] (2 superfamilies)
barrel, closed; n=6, S=10; greek-key
Superfamily b.44.1: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain [50465] (1 family) 
probably related to the second domain and its superfamiy by a circular permutation
Family b.44.1.1: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain [50466] (6 proteins) 
Protein Sulfate adenylate transferase subunit cysN/C, EF-Tu domain 3-like domain [141345] (1 species) 
Species Pseudomonas syringae pv. tomato [TaxId:323] [141346] (1 PDB entry) 
Domain d1zunb2: 1zun B:330-434 [125678]
Other proteins in same PDB: d1zuna1, d1zunb1, d1zunb3
complexed with ags, gdp, mg, na
Details for d1zunb2
PDB Entry: 1zun (more details), 2.7 Å
PDB Description: Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae
PDB Compounds: (B:) sulfate adenylate transferase, subunit 1/adenylylsulfate kinaseSCOP Domain Sequences for d1zunb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1zunb2 b.44.1.1 (B:330-434) Sulfate adenylate transferase subunit cysN/C, EF-Tu domain 3-like domain {Pseudomonas syringae pv. tomato [TaxId: 323]}
qvsdafdamlvwmaeepmlpgkkydikratsyvpgsiasithrvdvntleegpasslqln
eigrvkvsldapialdgyssnrttgafividrltngtvaagmiia
SCOP Domain Coordinates for d1zunb2:
Click to download the PDB-style file with coordinates for d1zunb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1zunb2:
- d1zunb2 is new in SCOP 1.73
- d1zunb2 appears in SCOP 1.75
- d1zunb2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D