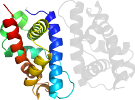
Lineage for d1ztdb2 (1ztd B:2-125)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.149: RNase III domain-like [69064] (1 superfamily)
core: 5 helices; one helix is surrounded by the others
Superfamily a.149.1: RNase III domain-like [69065] (3 families) 

Family a.149.1.2: PF0609-like [140675] (2 proteins)
specific to Thermococci; shares conserved carboxylic residues and similar dimerisation mode with the RNase III domain; contains integrated in the fold extra C-terminal helix, making it similar to the core fold of Ribosomal protein S7 (47972)
automatically mapped to Pfam PF11469
Protein automated matches [190862] (1 species)
not a true protein
Species Pyrococcus furiosus [TaxId:2261] [188202] (1 PDB entry)
automatically mapped to Pfam PF09010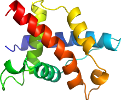
Domain d1ztdb2: 1ztd B:2-125 [125640]
Other proteins in same PDB: d1ztda1, d1ztda2, d1ztdb3
automated match to d1ztda1
Details for d1ztdb2
PDB Entry: 1ztd (more details), 2 Å
PDB Description: hypothetical protein pfu-631545-001 from pyrococcus furiosus
PDB Compounds: (B:) Hypothetical Protein Pfu-631545-001SCOPe Domain Sequences for d1ztdb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ztdb2 a.149.1.2 (B:2-125) automated matches {Pyrococcus furiosus [TaxId: 2261]}
eidkglakfgdslinflyslalteflgkptgdrvpnaslaialeltglsknlrrvdkhak
gdyaealiakawlmglisereaveiikknlypevldfskkkeaigralapllviiserly
ssqv
SCOPe Domain Coordinates for d1ztdb2:
Click to download the PDB-style file with coordinates for d1ztdb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ztdb2:
- d1ztdb2 first appeared in SCOP 1.73, called d1ztdb1
- d1ztdb2 appears in SCOPe 2.07

 View in 3D
View in 3D