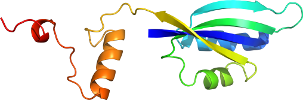
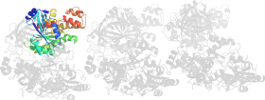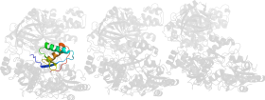Lineage for d1zm3a5 (1zm3 A:726-842)
- Root: SCOPe 2.04

Class d: Alpha and beta proteins (a+b) [53931] (380 folds) 
Fold d.58: Ferredoxin-like [54861] (59 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.11: EF-G C-terminal domain-like [54980] (4 families) 
Family d.58.11.0: automated matches [254210] (1 protein)
not a true family
Protein automated matches [254469] (2 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [255012] (4 PDB entries) 
Domain d1zm3a5: 1zm3 A:726-842 [125302]
Other proteins in same PDB: d1zm3a1, d1zm3a2, d1zm3a3, d1zm3b_, d1zm3c1, d1zm3c2, d1zm3c3, d1zm3d_, d1zm3e1, d1zm3e2, d1zm3e3, d1zm3f_
automated match to d1n0ua5
Details for d1zm3a5
PDB Entry: 1zm3 (more details), 3.07 Å
PDB Description: Structure of the apo eEF2-ETA complex
PDB Compounds: (A:) Elongation factor 2SCOPe Domain Sequences for d1zm3a5:
Sequence; same for both SEQRES and ATOM records: (download)
>d1zm3a5 d.58.11.0 (A:726-842) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
epvflveiqcpeqavggiysvlnkkrgqvvseeqrpgtplftvkaylpvnesfgftgelr
qatggqafpqmvfdhwstlgsdpldptskageivlaarkrhgmkeevpgwqeyydkl
SCOPe Domain Coordinates for d1zm3a5:
Click to download the PDB-style file with coordinates for d1zm3a5.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1zm3a5:
- d1zm3a5 first appeared in SCOP 1.73
- d1zm3a5 appears in SCOPe 2.03
- d1zm3a5 appears in SCOPe 2.05
- d1zm3a5 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from same chain: (mouse over for more information) d1zm3a1, d1zm3a2, d1zm3a3, d1zm3a4 |