Lineage for d1zm3a1 (1zm3 A:344-481)
- Root: SCOP 1.73

Class b: All beta proteins [48724] (165 folds) 
Fold b.43: Reductase/isomerase/elongation factor common domain [50412] (4 superfamilies)
barrel, closed; n=6, S=10; greek-key
Superfamily b.43.3: Translation proteins [50447] (5 families) 
Family b.43.3.1: Elongation factors [50448] (10 proteins) 
Protein Elongation factor 2 (eEF-2), domain II [82118] (1 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [82119] (13 PDB entries) 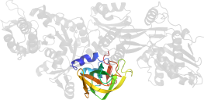
Domain d1zm3a1: 1zm3 A:344-481 [125298]
Other proteins in same PDB: d1zm3a2, d1zm3a3, d1zm3a4, d1zm3a5, d1zm3b1, d1zm3c2, d1zm3c3, d1zm3c4, d1zm3c5, d1zm3d1, d1zm3e2, d1zm3e3, d1zm3e4, d1zm3e5, d1zm3f1
automatically matched to d1n0ua1
complexed with dde
Details for d1zm3a1
PDB Entry: 1zm3 (more details), 3.07 Å
PDB Description: Structure of the apo eEF2-ETA complex
PDB Compounds: (A:) Elongation factor 2SCOP Domain Sequences for d1zm3a1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1zm3a1 b.43.3.1 (A:344-481) Elongation factor 2 (eEF-2), domain II {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
spvtaqayraeqlyegpaddanciaikncdpkadlmlyvskmvptsdkgrfyafgrvfag
tvksgqkvriqgpnyvpgkkddlfikaiqrvvlmmgrfvepiddcpagniiglvgidqfl
lktgtlttsetahnmkvm
SCOP Domain Coordinates for d1zm3a1:
Click to download the PDB-style file with coordinates for d1zm3a1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1zm3a1:
- d1zm3a1 is new in SCOP 1.73
- d1zm3a1 appears in SCOP 1.75
- d1zm3a1 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from same chain: (mouse over for more information) d1zm3a2, d1zm3a3, d1zm3a4, d1zm3a5 |