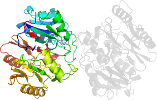
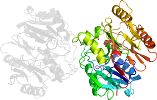Lineage for d1zbra1 (1zbr A:3-341)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.126: Pentein, beta/alpha-propeller [55908] (1 superfamily)
duplication: composed of 5 alpha-beta(2)-alpha-beta units arranged around pseudo fivefold axis
Superfamily d.126.1: Pentein [55909] (8 families) 

Family d.126.1.6: Porphyromonas-type peptidylarginine deiminase [111155] (3 proteins)
Pfam PF04371; functionally related to the amidinotransferase, similar active site
Protein Putative peptidyl-arginine deiminase [111158] (3 species) 
Species Porphyromonas gingivalis [TaxId:837] [143808] (1 PDB entry)
Uniprot Q7MXM8 3-341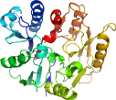
Domain d1zbra1: 1zbr A:3-341 [124868]
Other proteins in same PDB: d1zbrb_
Details for d1zbra1
PDB Entry: 1zbr (more details), 2.6 Å
PDB Description: crystal structure of the putative arginine deiminase from porphyromonas gingivalis, northeast structural genomics target pgr3
PDB Compounds: (A:) conserved hypothetical proteinSCOPe Domain Sequences for d1zbra1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1zbra1 d.126.1.6 (A:3-341) Putative peptidyl-arginine deiminase {Porphyromonas gingivalis [TaxId: 837]}
krlflpewapqeavqltwphdrtdwaymldevetcfvriatailrherlivvcpdrkrvf
gllppelhhrlycfelpsndtwardhggislladgrpmiadfafngwgmkfaahhdnlit
rrlhalglfaegvtldnrlafvleggaletdgegtllttdsclfepnrnaglsrtaiidt
lkeslgvsrvlslrhgalagddtdghidtlarfvdtrtivyvrsedpsdehysdltameq
elkelrrpdgqpyrlvplpmaealydgadrlpatyanfliingavlvptydshldavals
vmqglfpdrevigidcrplvkqhgslhcvtmqypqgfir
SCOPe Domain Coordinates for d1zbra1:
Click to download the PDB-style file with coordinates for d1zbra1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1zbra1:
- d1zbra1 first appeared in SCOP 1.73
- d1zbra1 appears in SCOPe 2.06
- d1zbra1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D